A deep dive into the dark regions of the human genome
deCODE scientists pull insights from the first 150,000 whole genomes in the UK Biobank
Happy Friday! My pick for this week’s “From the Twitter archives” is a thread I wrote in Nov 2021 on a flagship paper from the deCODE genetics on the first release of 150k UK Biobank whole genome sequences. I wrote it when the work was preprinted. The paper was published in Nature in July 2022 and was widely celebrated. You might also want to check out deCODE’s video press release where Kari Stefansson, the last author, interviews Bjarni Halldorsson, the first author (deCODE publishes a press release on the news page of their website after every major publication, and recently their PR people have gotten more creative. Every post now has a professionally taken photograph of Kari and the lead author posed in interesting ways! I never miss checking out their press releases)
From the Twitter archives
In this milestone paper in human genetics, deCODE scientists (the very people who pioneered the art of population-scale whole genome sequencing) illuminate the dark side of the human genome in >150,000 individuals.
We've been pondering over the question of what WGS will reveal that whole exome sequencing (WES) didn't, since the day UKB announced the release date of 200k whole genomes.
Well, deCODE scientists have beautifully shown in this paper that there are tons of fascinating things one can do using whole genomes, things that cannot be done using whole exomes.
Too many fascinating findings to fit in a single thread. But few of them deserve a highlight.
WES is not truly "whole exome" sequencing.
One that might come as a surprise for at least a few of us is that WGS informs not only about the non-coding genome but also about parts (~10%) of the coding genome that are usually missed by WES.
Though it's called "whole exomes", WES informs little about the parts of the exons—5' and 3' UTRs—that are transcribed but not translated. WGS captures it all.
As an example, the authors highlight a rare 5'UTR splice acceptor variant in TAC3 (Tachykinin 3) present in 370 women in UKB that delays menarche by 11 months! (WES missed it). Complete loss of TAC3 (or its receptor TACR3) causes hypogonadotropic hypogonadism, an autosomal recessive condition. Now we know partial loss is not without phenotypic consequences. TAC3 is another beautiful addition to the growing list of reports of heterozygous effects for presumed recessive genes.
Rare non-coding variants with large effects.
Rare non-coding variants with large effects are one main thing about WGS that excites many of us, and the authors didn't disappoint us. They have dug out many fascinating non-coding variants.
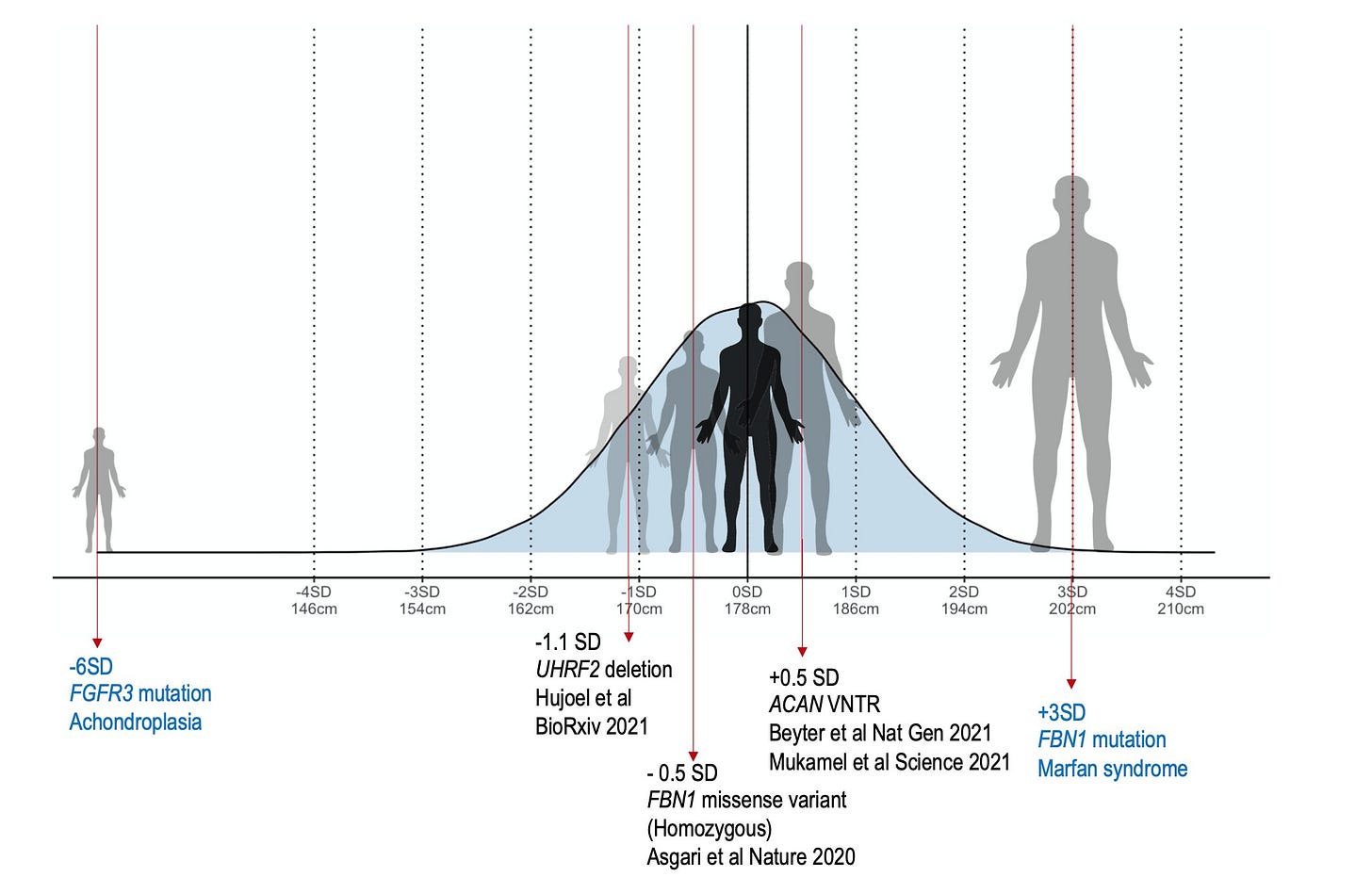
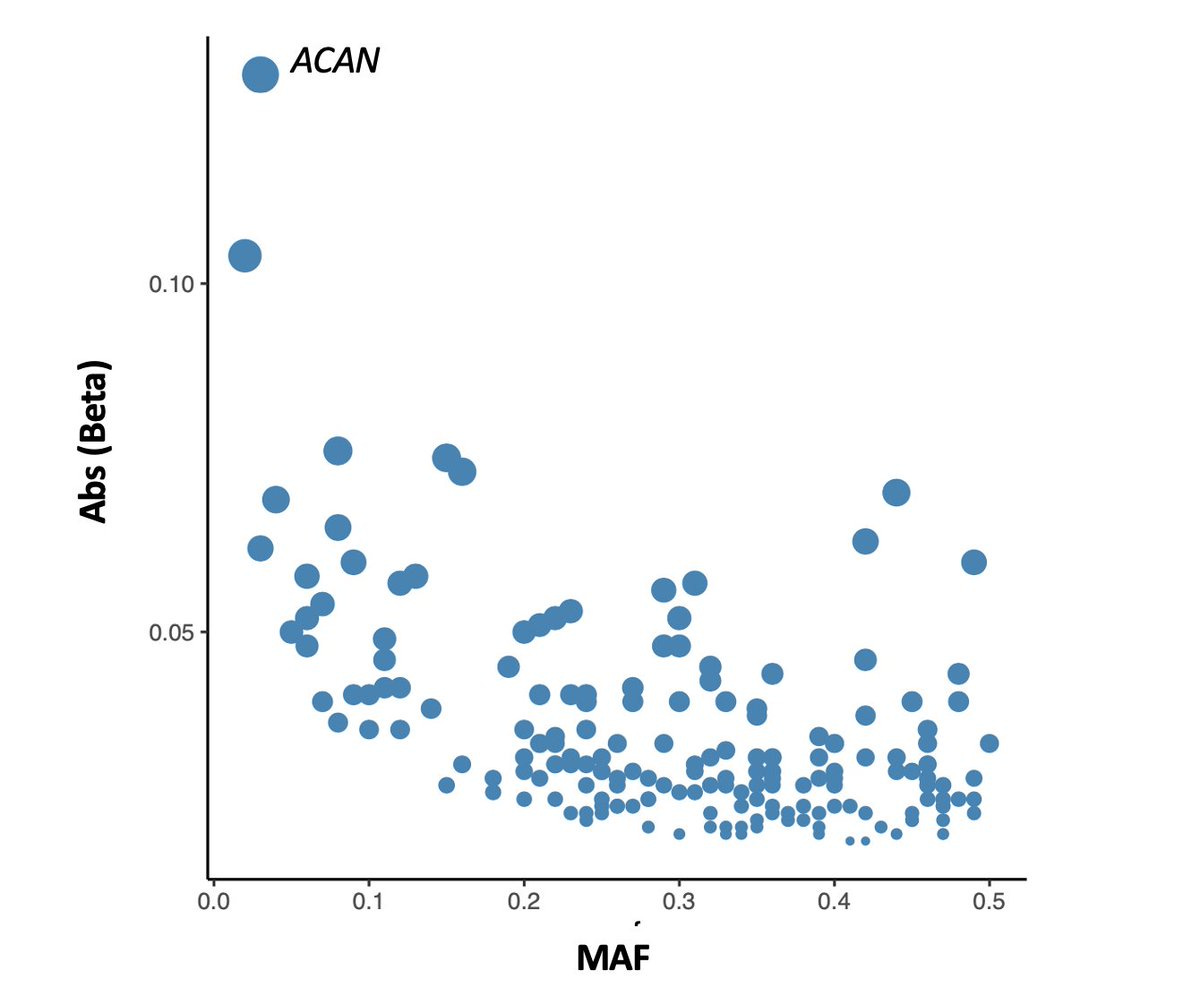
For example, an SNV in the promoter of GHRH (growth hormone-releasing hormone) reduces height by ~3cm (0.32 sd). This is larger than the largest effect size identified in height GWAS (as you can see in the plot below I made a year ago).
Note, this is still smaller than the maximum effect size of the ACAN VNTR, but slightly larger than the heterozygous effect size (2.2cm) of Peruvians specific FBN1 missense variant.
It's exciting to imagine many more such variants are sitting quietly in the non-coding genome within critical regulatory elements waiting to be discovered.
Footprints of natural selection in the non-coding genome.
After studying millions of exomes, now we have a pretty good understanding of how natural selection shaped the coding variations. But we are still scratching the surface when it comes to non-coding variants.
Here the authors leveraged the ~150k whole genomes and identified regions of the genome that are depleted of mutations, like the regions of the plane below (an iconic figure widely shared on the internet to illustrate survival bias) that are not hit by bullets.
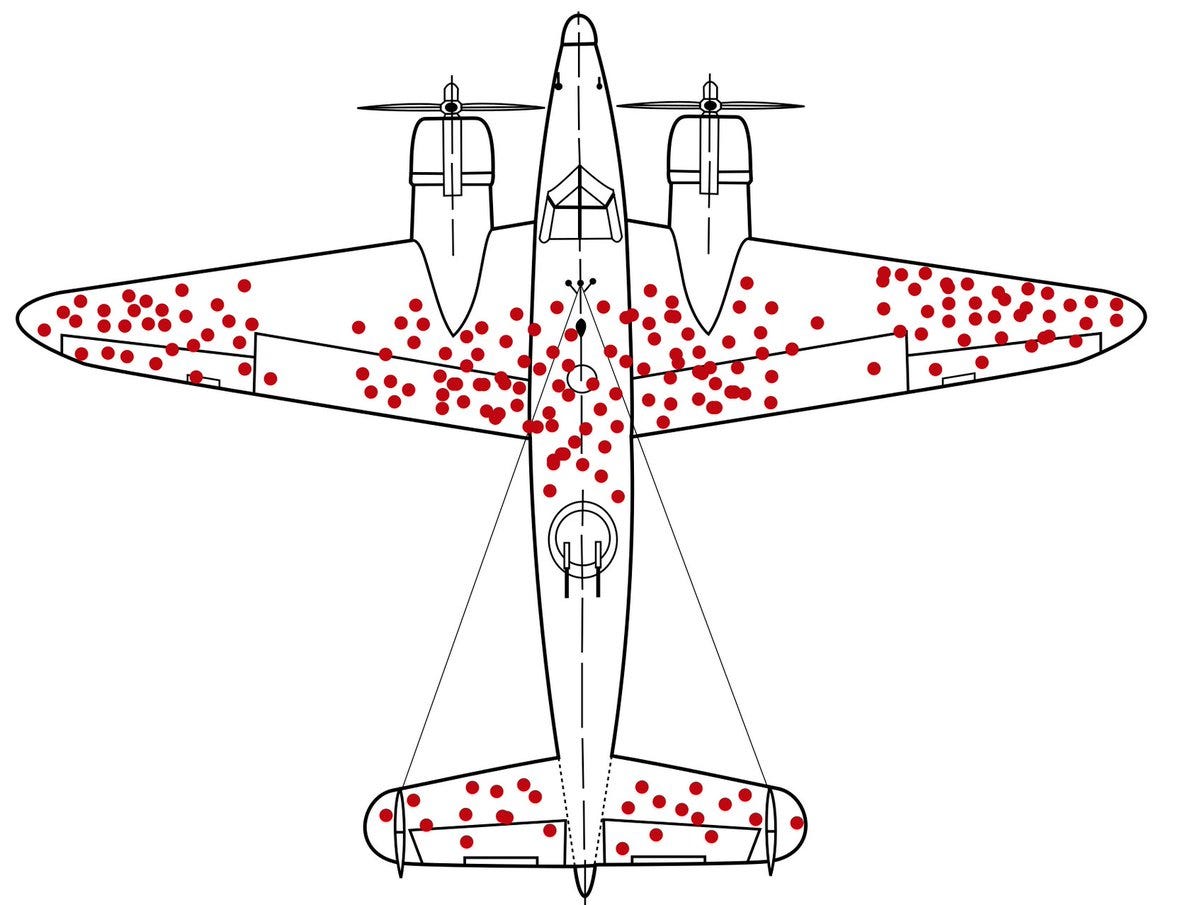
Among the top 1% most mutation-depleted regions are exons (obviously), splice regions, UTRs, and genes upstream and downstream. And half of these most mutation-depleted regions weren't conserved between humans and other species. Such regions are where negative selection has happened exclusively in humans. That's super fascinating!
The hunt for structural variants (beasts hiding in plain sight)
When someone says WGS, many of us hear structural variants--the single most important utility of WGS.
Of course, the authors didn't disappoint us. They have discovered many of those beautiful beasts:
~14kb deletion of the first exon of PCSK9 (b=-1.22 sd)
~4kb deletion of ALB promoter (b=-1.5 sd)
a ~16kb deletion that fully removes 2 exons of GCSH (b=1.45 sd)
One specific aspect of these SVs that I particularly love is many of them masquerade as innocent LD variants in the GWAS.
The authors found a 754bp deletion that removes exon 6 of NMRK2 leading to an earlier age at menopause. It turns out it's the causal variant at a previously identified GWAS locus for age at menopause (plot from open targets)
Next-generation high-resolution imputation panels
deCODE scientists have mastered the art of deriving haplotypes from WGS and imputing rare variants from array data. As I expected they didn't miss this opportunity to show off their skills. The authors created an imputation panel using which we can now accurately impute >98.5% of variants with MAF down to 0.1% and 65.8% of variants with MAF down to 0.001%(!)
But the most important thing is this new imputation panel extra-ordinarily benefits African and South-Asian populations. People doing GWAS in South-Asian and African populations should make use of this reference panel.
The authors highlight many striking examples of clinically important variants that we can now impute from array data without the need for sequencing.
E.g. a pathogenic 5' UTR beta-thalassemia variant in HBB, a stop gain variant in PCSK9 common in Africans. They even found one African individual who is homozygous (i.e. a human knockout for PCSK9).
WGS offers a deeper peek into our ancestors.
For more info on this, please see Kristján's insightful Twitter thread.
There are many more fascinating findings packed in the paper. This is a must-read. Many congrats to the whole deCODE team for this phenomenal work.






Thank you for writing about the fascinating power our genome has in regulating ocmplex traits. It is fascinating to see the effect sizes! 🤩