A master class on human genetics and drug development
Using allelic series to infer drug efficacy and adverse effects
Happy Friday! It was a busy week for me. I was trying to complete a post on allelic series that I started writing a week ago. But I couldn’t find time. I’ll get it done this weekend. As I’ve been thinking about allelic series a lot this week, I felt a past Twitter thread on the same topic would be a good choice for this week’s From the Twitter archives post.
From the Twitter archives
During the 2021 American Society of Human Genetics (ASHG) conference, I listened to a talk by Robert Plenge (the head of the research at Bristol-Myers Squibb) on how genetics findings can aid in drug discovery. I think it's one of the best talks on this topic I've ever listened to. Here I list some snippets of this talk.
One of the best lines of genetic evidence to support drug discovery is allelic series—multiple variants at a locus independently influencing a phenotype (assuming a single causal gene).
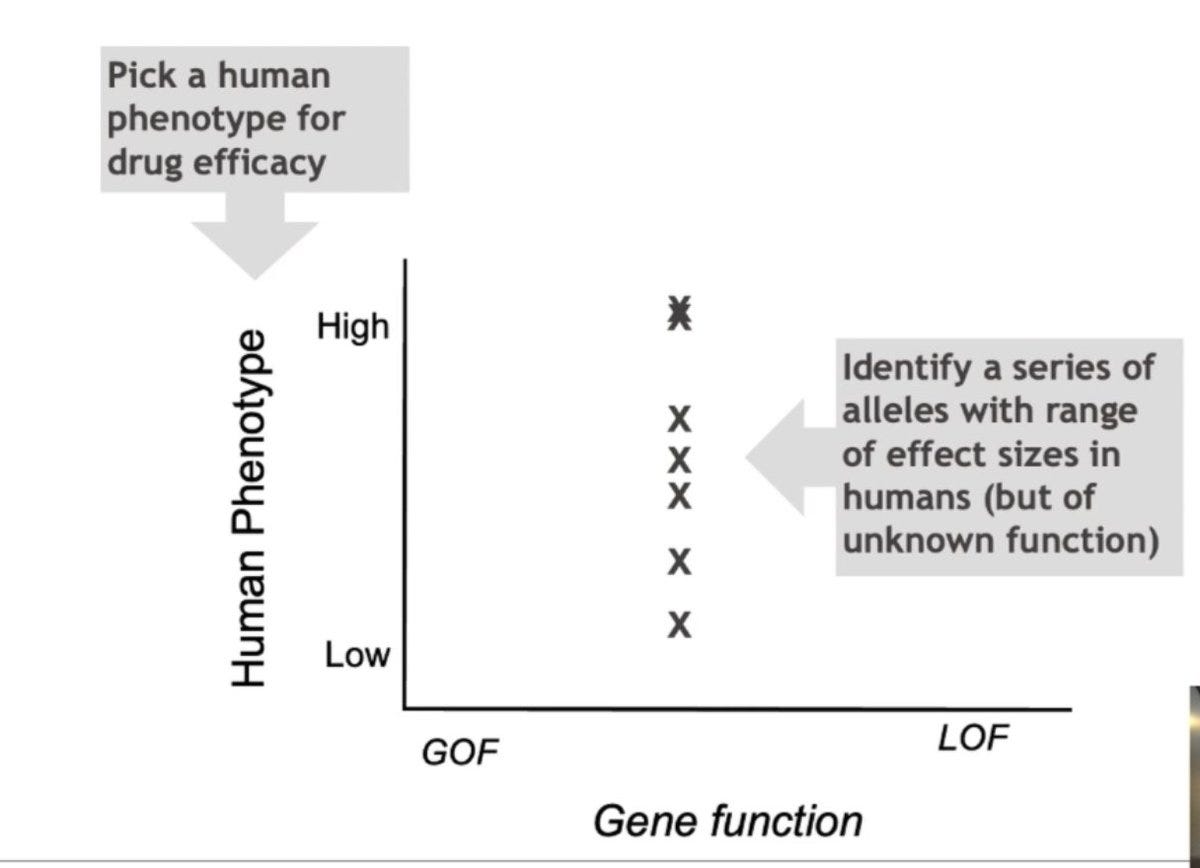
If you want to study genetic evidence for a drug candidate for a disease (e.g.protein inhibitor), first identify all the possible alleles that are significantly associated with the disease at varying effect sizes.
Identify the molecular mechanisms of the alleles i.e. if the allele leads to loss of function (LOF) or gain of function (GOF) or if it slightly increases or decreases the gene expression.
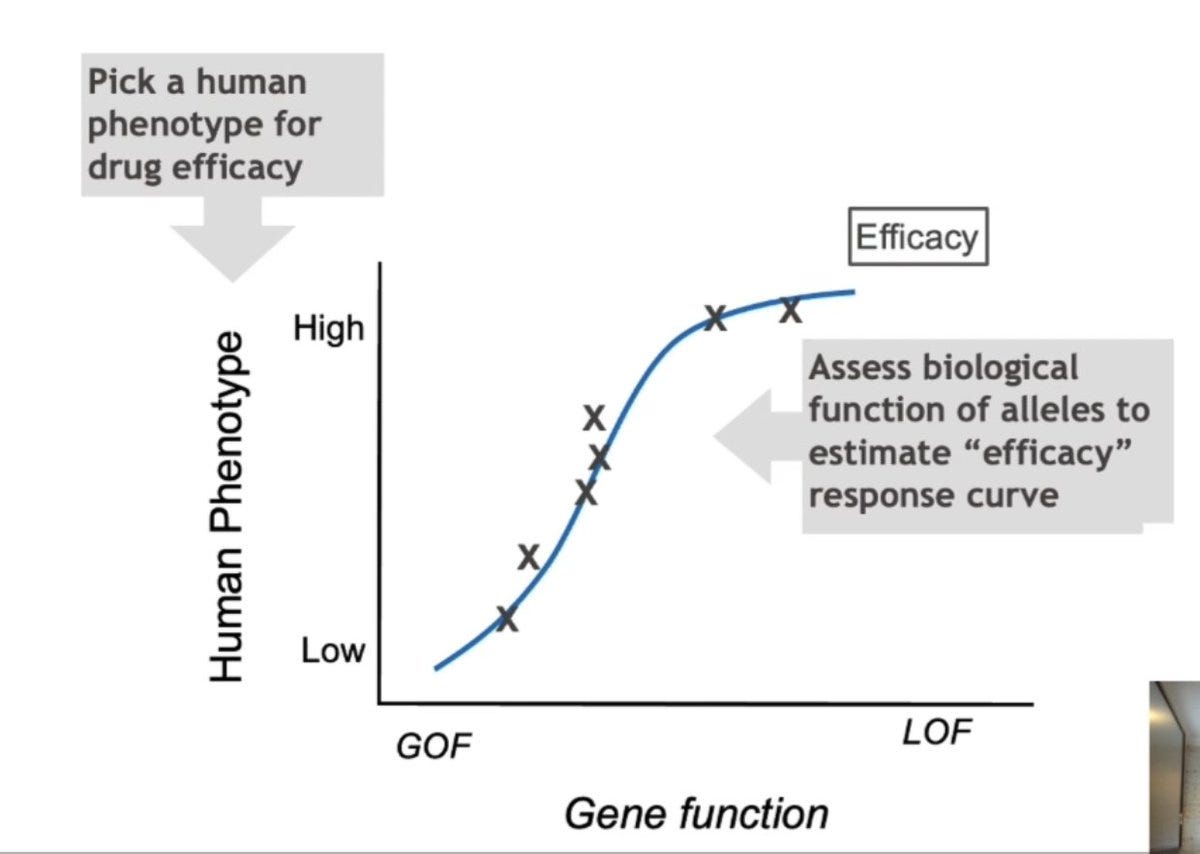
When you plot the mechanism on the X-axis and effect size on the Y-axis, what you get is a genetic dose-response relationship, which is a surrogate for the drug-dose response relationship.
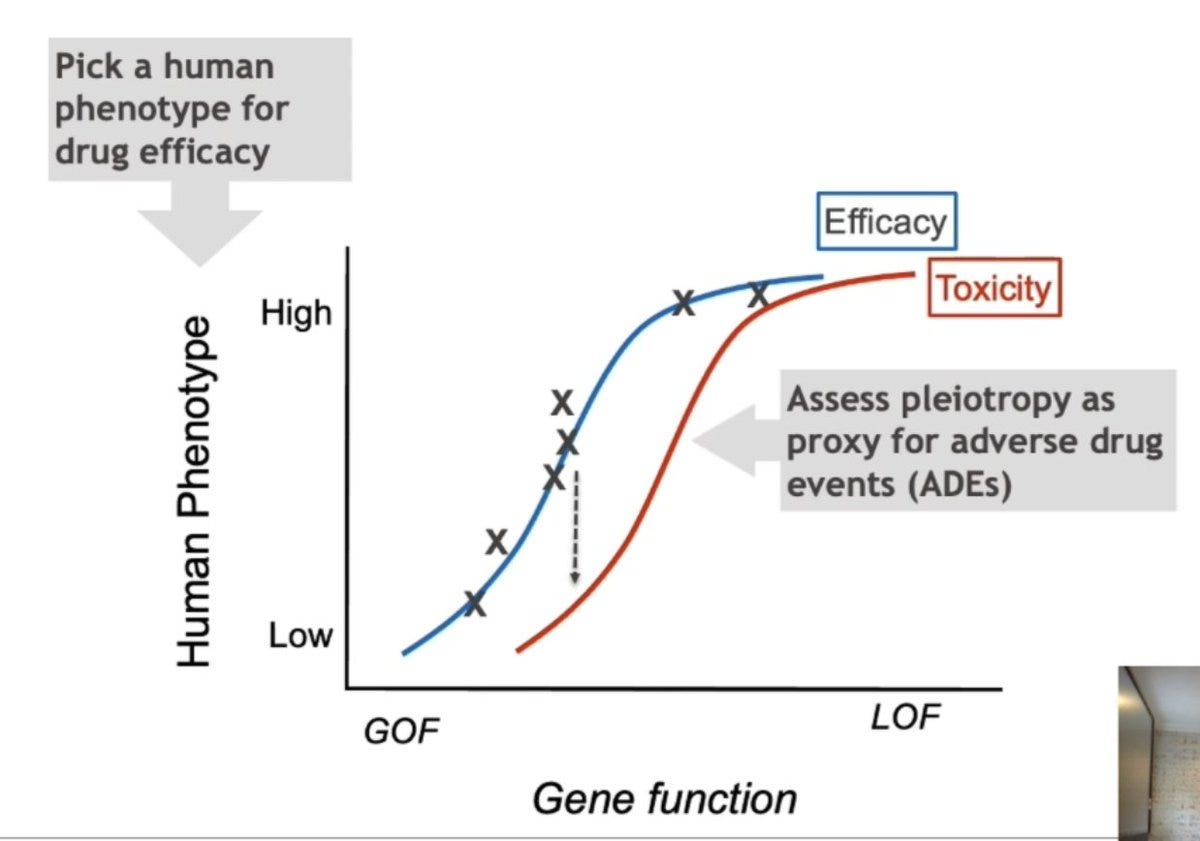
This not only informs the efficacy of the drug, but when visualized for a pleiotropic phenotype (that might be a potential side effect of the drug), you also get a dose-toxicity response relationship.
If you can succeed in getting this data for your drug candidate, then the probability of success in clinical trials is significantly high, which has now been shown retrospectively for many drug candidates.
Robert illustrated the above workflow using the TYK2 inhibitor that his company has developed for the treatment of psoriasis as an example.
GWASs show that coding variants that reduce TYK2 gene function are protective for auto-immune diseases like rheumatoid arthritis and psoriasis. But at the same time, pathogenic mutations in TYK2 leading to complete loss of function results in immunodeficiency. If so, will an inhibitor of TYK2 treat auto-immunity without causing immunodeficiency and increased risk for infections?
Using genetics alone, Robert and his colleagues confirmed that 50 to 80% of TYK2 inhibition can be achieved without potentially increasing infection risk. Based on that evidence, they designed a small molecule TYK2 inhibitor that has shown promising results in phase II and III clinical trials for psoriasis without causing major adverse events.
Thanks to Robert for this master class. For further reading here is a blog post by Robert Plenge on this same topic.
P.S. Robert replied to my thread with a link to the video recording of his presentation, which is still active.




Interesting, thanks Veera. With respect to the specific data on TYK2 and infection, do we not just think this is a power issue. Infection is rare, and I would be worried about the power of these approaches in the linked paper in PLOS Med. Although I buy the principle!