A peek into the enigmatic universe of noncoding mutations
Studying polydactyly mutations in humans and mice uncovers a novel noncoding mechanism that governs tissue-specific gene expression
Hope your weekend is going great. Here is a new story to add to your weekend science reads. Enjoy!
A new paper in Nature by Emma Farley and colleagues from the University of California reminds us again that we have hardly scratched the surface of exploring the enigmatic universe of noncoding variants.
As I often say on Twitter, the most beautiful, in-depth biological insights into the human genome often come not from sequencing hundreds of thousands of individuals from the general population but from a handful of patients with extreme phenotypes that clinicians encounter in the clinics. This is true for both coding and noncoding variants, but is much more relevant for noncoding variants than coding variants, given the recent excitement around the release of half a million genomes of the UK Biobank participants.
Probing the mechanisms underlying some noncoding enhancer mutations that cause polydactyly in humans and mice, the authors uncover a surprising mechanism (that might have been perfected over millions of years of evolution) through which enhancers control the tissue-specific expression of their target genes.
If asked to guess about how binding of certain transcription factors (TFs) to unique nucleotide sequence patterns (motifs) within enhancers results in tissue-specific (and/or developmental stage-specific) expression, we would imagine that the TFs bind to motifs with high affinity and any mutation that scramble the motifs would result in TFs failing to bind to the enhancers resulting in aberrant gene expression.
Well, it turns out some enhancers like ZRS (mutations in which cause polydactyly) were designed by evolution to act in a manner exactly opposite to what we would imagine.
ZRS is highly conserved across vertebrates with near complete sequence identity between humans and mice (there were instances where the same nucleotide change has caused the same phenotype in humans and mice). It controls a gene called SHH (Shh in mice) that encodes sonic hedgehog protein that plays a critical role in limb development. Mutations in the ZRS enhancer cause polydactyly, which has been observed even in cats. Here is a polydactyly Hemingway cat.
Previous studies have shown that the ZRS mutations are a gain of function in nature and cause polydactyly through ectopic expression of SHH in the developing limb bud. But how exactly the mutations cause ectopic expression remained a mystery.
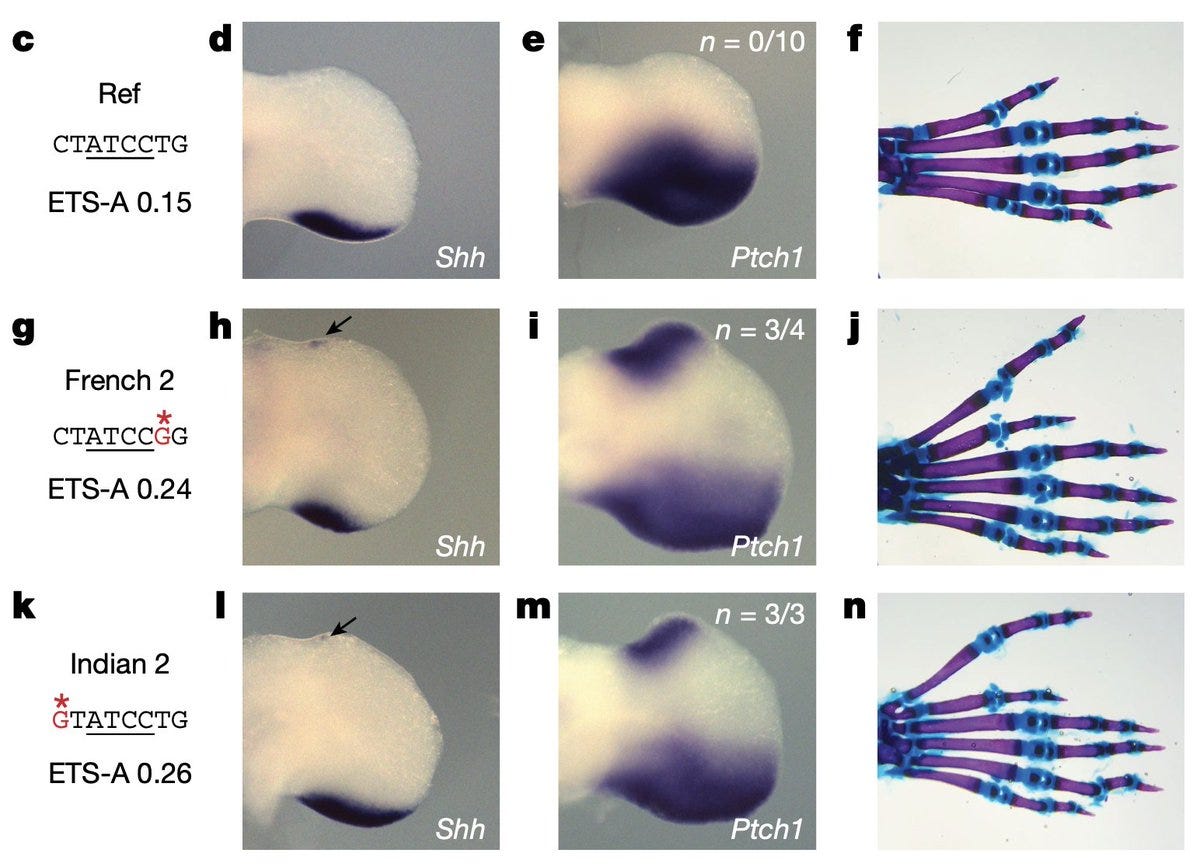
In the current work, the authors find that the ZRS enhancers employ a regulatory phenomenon (previously observed in invertebrates) where enhancers maintain tissue specificity by using multiple, redundant low-affinity binding sites for transcription factors. The binding affinity between enhancer motifs and transcription factors is kept at extremely low levels like 15% of the maximum possible affinity.
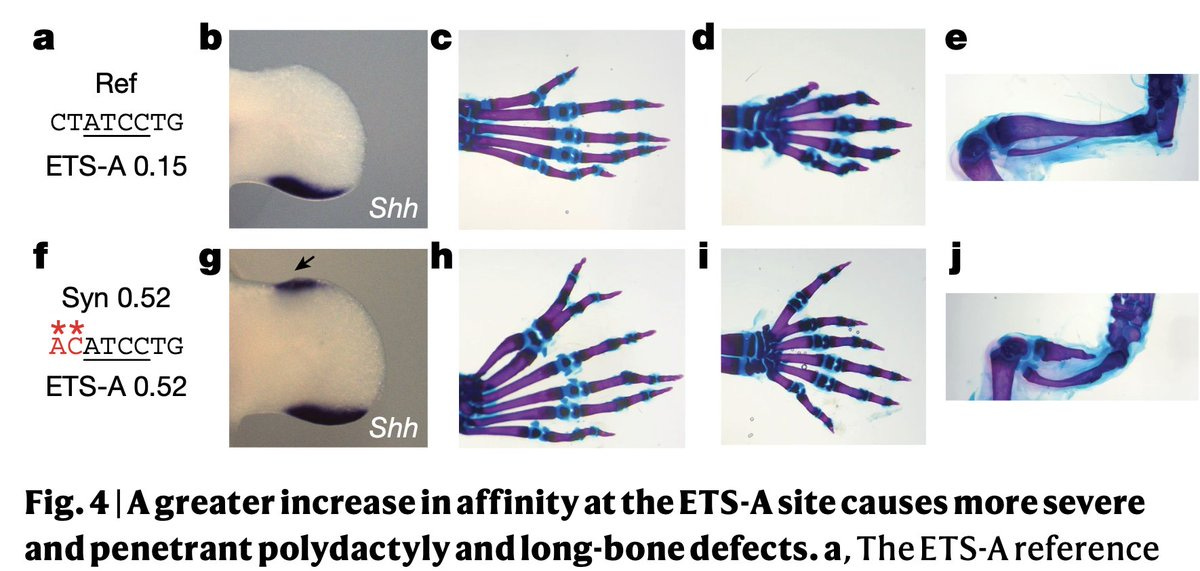
The sub-optimal and transient binding of TFs to ZRS during organ development ensures that its target genes are expressed only within a critical zone of the developing limb bud. Mutations that increase the affinity of ZRS to their TFs result in spillover of expression resulting in the development of extra fingers. The image below shows the ectopic expression of Shh and its downstream target Ptch1 in the background of polydactyly mutation.
The effect of gain of function mutations is dose-dependent. The higher the affinity, the more severe the polydactyly. The consequences can be so severe that mutations sometimes result in an extra whole organ, for example, two beating hearts (!) previously observed in sea squirts.
What’s more fascinating is the redundancy of such enhancer elements. By placing the control of gene expression under multiple redundant enhancer elements with low-affinity binding sites, the evolution has tried to make the system error-proof as the loss of one enhancer site will be buffered by the other. But fascinatingly, the master design came with a loophole—vulnerability towards the gain of function mutations. Perhaps, having an extra finger is a small price the organisms had to pay to avoid losing the limb altogether.
Another great insight into the noncoding variants that we learn from this amazing work is the challenge behind designing in vitro experiments like massively parallel reporter assay (MPRA) to identify mutations in enhancers that follow the low-affinity binding principle. We often focus on the variants that produce the maximal change in the gene expression in cellular assays to prioritize functional variants. Such an approach will miss variants like the ones in ZRS that cause only subtle changes in the gene expression but have a massive effect on the phenotypes.
Overall, it’s a truly amazing work offering novel insights into the mechanisms of noncoding mutations.
Thanks for reading! If you loved the story, consider sharing it with a friend. Also, do you know that you can send your feedback by simply replying to this email? Do send any feedback or suggestions that would help me improve the GWAS stories newsletter.
-Veera