A receptor meets its ligand through human genetics
Happy Friday! Many things at work have been keeping me busy for the past many days. Still, I don’t miss taking a peek into my Twitter feeds now and then to catch up on the latest news. Matt Nelson, who’s famously known for his 2015 paper on the impact of human genetics on success of drug programs, has published a new paper in Nature. In this updated analysis, led by Eric Vallabh Minikel from Broad Institute, the authors report that the probability of success (measured in terms of clearing regulatory approval and entering market) is now 2.6-fold higher when a drug candidate has human genetics support. It’s an interesting topic, which I’ll try to discuss more in a future post.
A few days ago, an incredible work was preprinted in the medRxiv by a large team of scientists from across the globe, led by Nicky Whiffin from Oxford University. Studying whole genomes of undiagnosed rare disease patients from the Genomics England, the authors stumbled on a mind-blowing discovery: a recurrent de novo non-protein coding variant within a highly conserved region of a small nuclear RNA gene, RNU4-2, explains ~0.5% of all undiagnosed neurodevelopmental disorders (NDDs) in the Genomics England cohort. That number is very big(!) for a de novo variant. Probably the largest ever known to date. This is going to solve the genetic diagnosis of thousands of children across the world, whose parents are probably waiting for years, seeing doctor after doctor, trying to find what genetic mutation is responsible for their children’s condition. I’ll discuss more on this preprint in a separate future post.
Now, coming back to our topic for the day, this week I wanted to share an old Twitter story of how human genetics and proteomics have led to a fascinating discovery of a receptor-ligand pair. When large scale pQTL studies gained momentum two years ago, thanks to new technology platforms like Olink and SomaScan, I wondered what kind of discoveries would be made. One thing that fascinated me is that pQTLs can illuminate receptor-ligand pairs. Genome-wide associations of protein levels in the blood typically reveal genetic signals in cis for the proteins under investigation. They also often reveal signals in trans for their interacting partners. Almost always, such receptor-ligand pairs were already known, which made me wonder if a novel receptor-ligand pair had ever been discovered using a pQTL study. One day, I stumbled upon one such example.
From the Twitter archives

A beautiful story of how human population genetic and proteomic studies have led to the discovery of SVEP1 as the natural ligand of the receptor PEAR1.
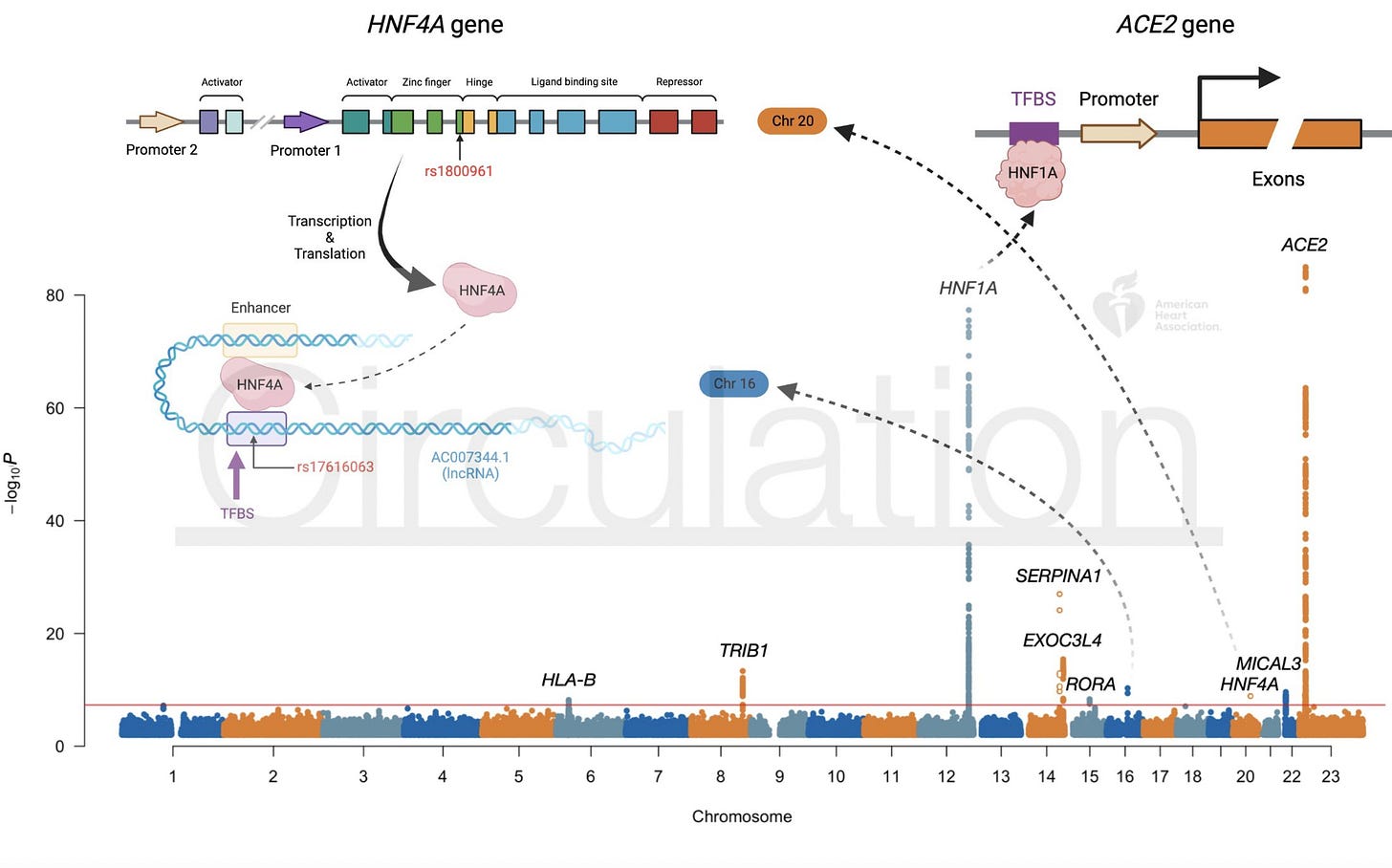
I am a big fan of GWAS of molecular traits as they often reveal clear and interpretable cis and trans signals reiterating the known biology of the trait. For example, a GWAS of a protein will reveal genetic variants that directly modify the protein expression in cis and also, genetic variants that indirectly modify the protein expression in trans. For example, below is a Manhattan plot from a GWAS of blood ACE2 levels showing a signal near ACE2 in cis and many signals in trans, including HNF1A and HNF4A, which are transcription factors that regulate ACE2 expression.
If the cis and trans pQTL signals can reveal known molecular interactions/connections (receptor-ligand, protein-TF), then, they should also be able to reveal previously unknown molecular interactions.
SVEP1-PEAR1 is one such rare example where a receptor-ligand discovery has been made via human proteomics (and also genomic) studies. Just for this fact, the paper by Elenbaas et al. will serve as an important reference in proteomics literature.
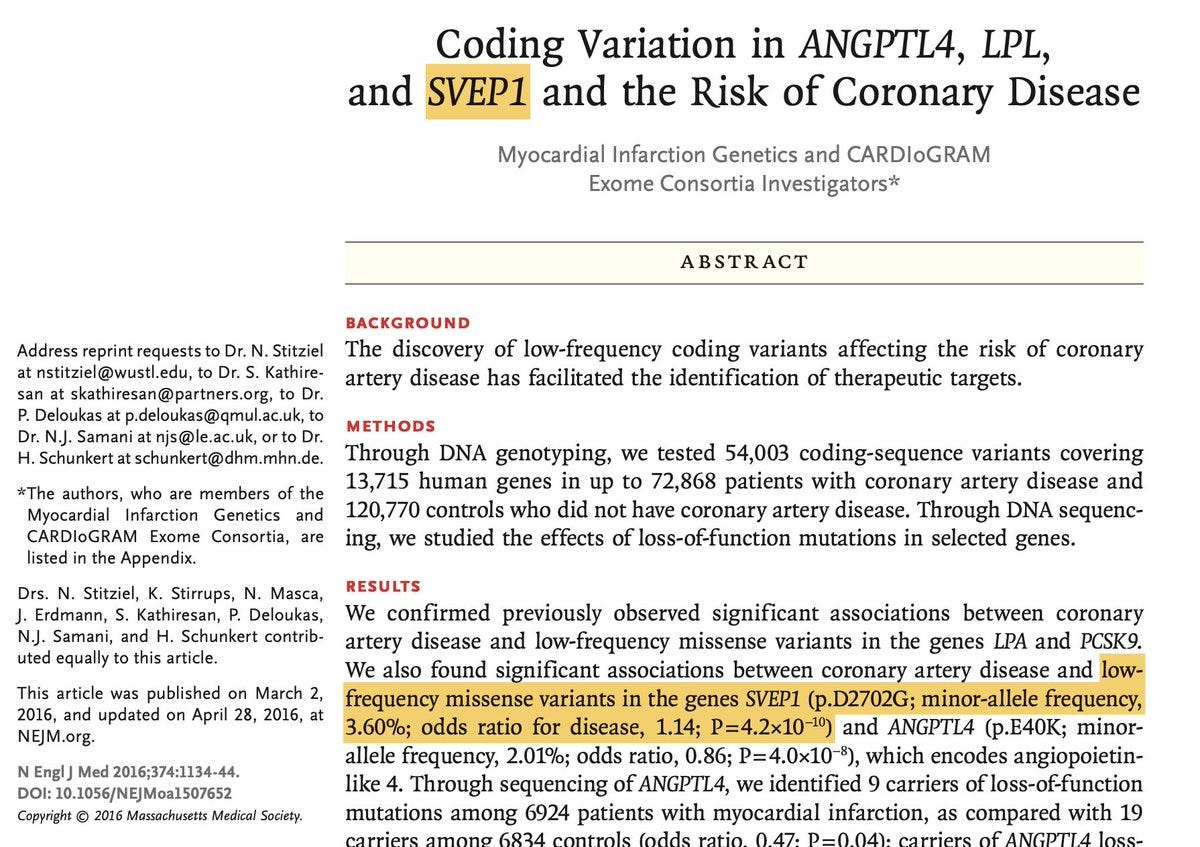
The background story of GWASs identifying disease associations for SVEP1 and PEAR1 in parallel without any knowledge of the secret affair between the two is fascinating. The story begins with the discovery of an association between a missense variant in SVEP1 (p.D2702G) and coronary artery disease (CAD) that appeared to be mediated not via the classical risk factor, lipids, but via something else, a novel mechanism.
SVEP1 codes for ‘sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing protein 1’ (that’s a mouthful name), which is an extracellular matrix protein. The authors of NEJM study later followed up the association via functional work to understand more about the role of SVEP1 in CAD. They found SVEP1 is expressed in vascular smooth muscle cells and acts as a proatherogenic factor, making it a potential target. Meanwhile, other studies linked SVEP1 genetic variants with a range of diseases including blood pressure, type 2 diabetes, septic shock and glaucoma.
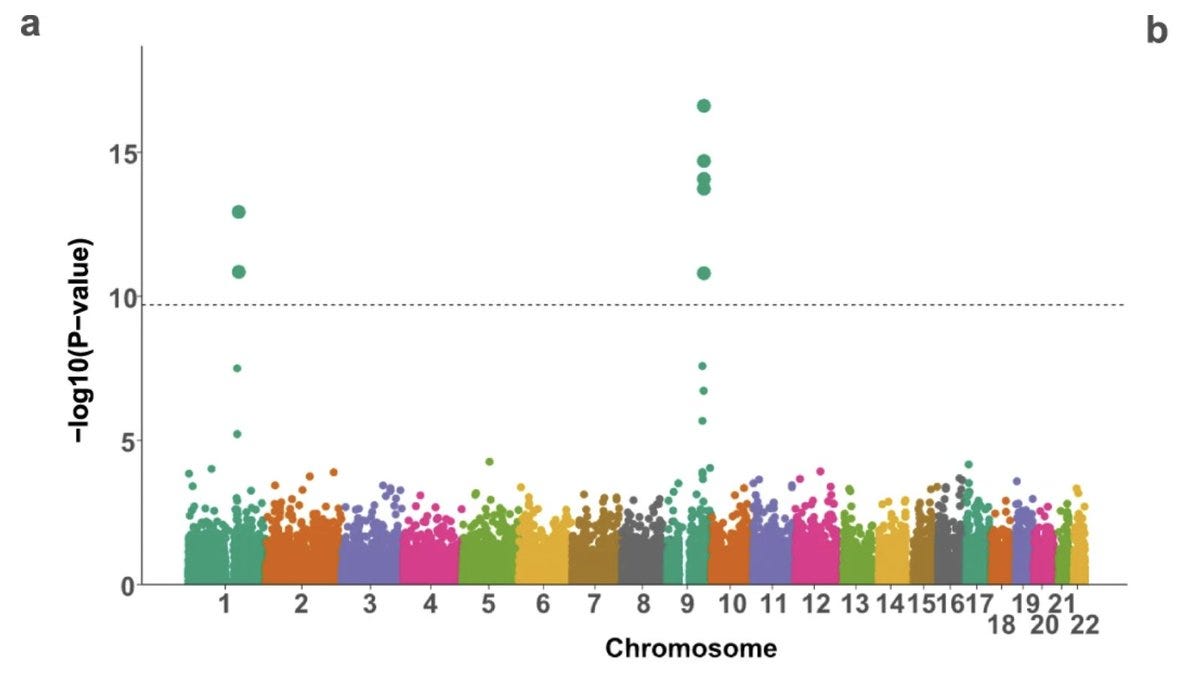
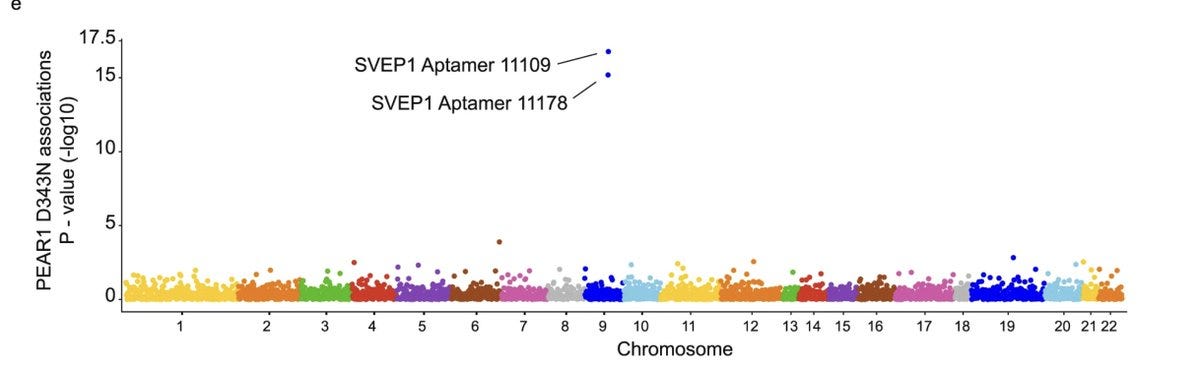
As the proteomics started catching up, the genetic link between SVEP1 and CAD became stronger from Mendelian Randomization analysis using SVEP1 cis pQTLs as genetic instruments. For example, an Icelandic study mapped the genetics of ~5k proteins in ~5k individuals, in which the authors highlighted the SVEP1 associations among others. The GWAS of SVEP1 levels in blood revealed two hits, one in cis (near SVEP1) and the other in trans (near PEAR1). The authors focussed only on the cis signal and ignored the trans signal (they likely had no idea at the time that the trans signal was actually pointing to the very receptor that SVEP1 binds to).
While SVEP1 associations were evolving in one world, in a parallel world, scientists were learning about PEAR1 through its strong genetic associations with platelet aggregation response (an important biological process in relation to atherosclerotic plaque formation & rupture). PEAR1 association with platelet response to agonists such as ADP is so strong it was found in an early study in mere 500 individuals published in 2009.
Unlike cis signals, trans signals will have relatively small effect sizes and so will require large sample sizes for discovery. The trans signal at SVEP1 for platelet aggregation appeared only in 2021 in a pQTL study of 3.8k individuals (that too only via gene based test).
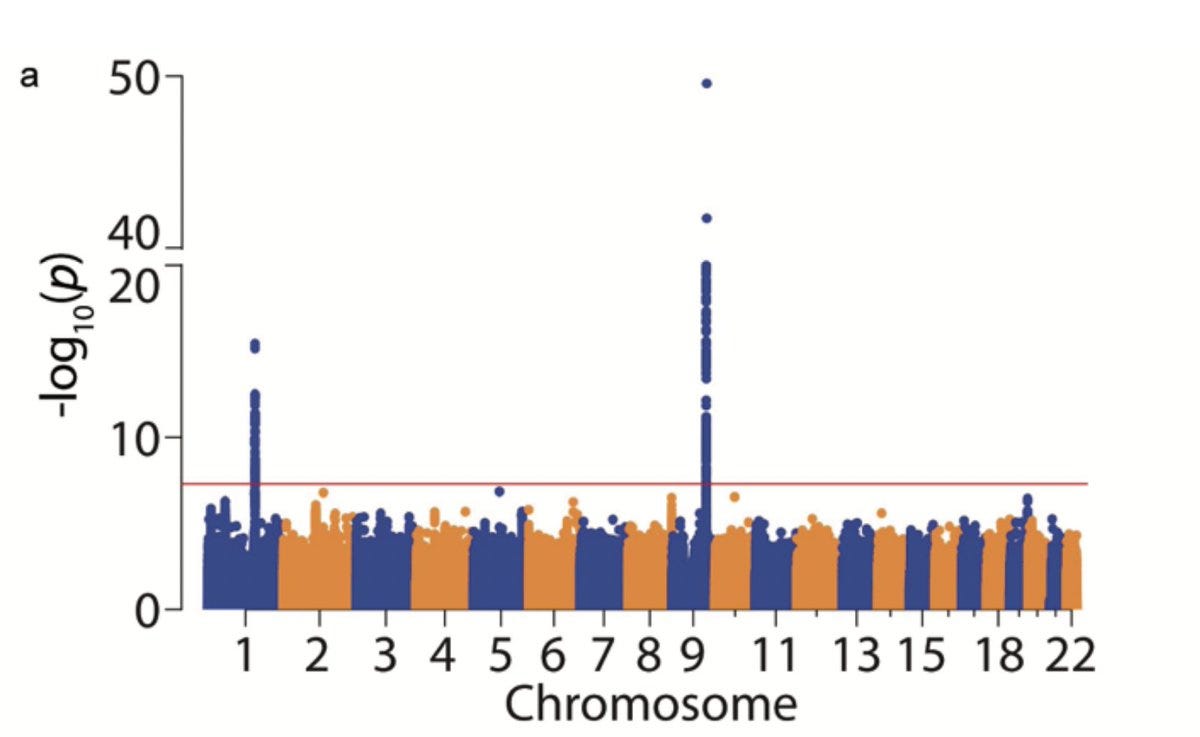
In the current paper, Elenbaas et al. connected all the dots, guessed that SVEP1 and PEAR1 are receptor-ligand pairs, and went on to prove the same using a series of proteomics, genomics and functional experiments. The authors reproduced the strong cis signal at SVEP1 and trans signal at PEAR1 using the pQTL data from the INTERVAL study.
Remarkably, the trans association in Chromosome 1 with PEAR1 missense variant was seen specifically with SVEP1 and not with any of the other 2994 proteins assayed in the INTERVAL study.
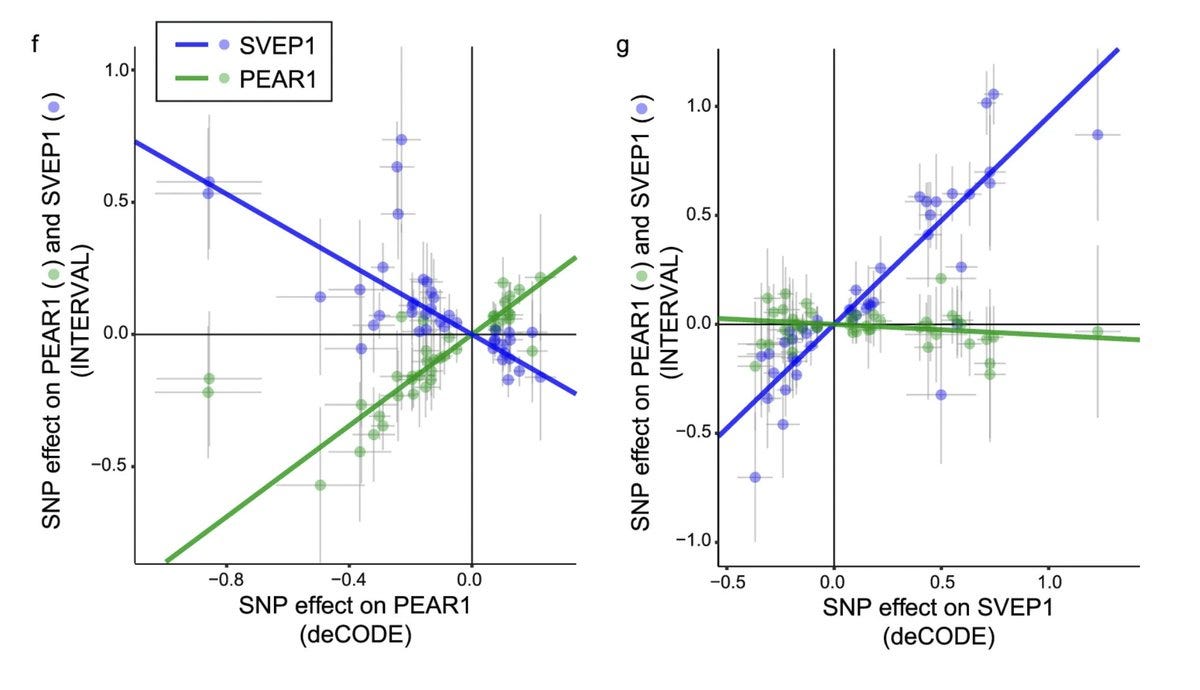
Based on pQTL results from an independent study (by deCODE Genetics) the authors show strong inverse relationship between PEAR1 and SVEP1 and further show that the causality direction is from receptor (PEAR1) to ligand (SVEP1) (i.e. increase in receptor decreases ligand and vice versa).
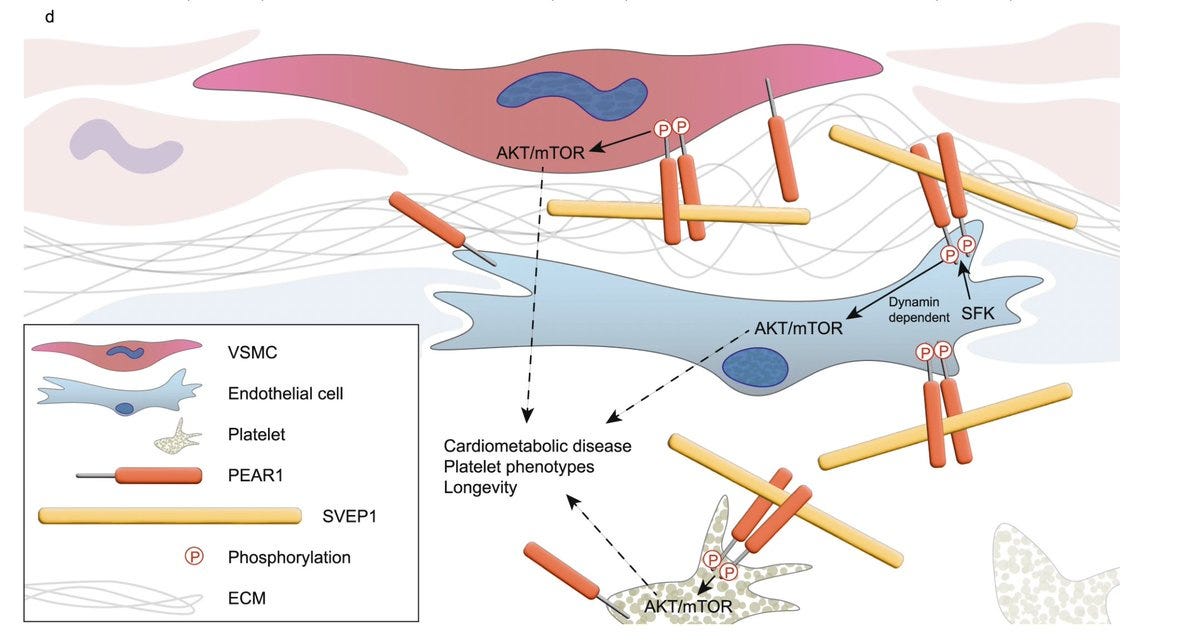
Through molecular assay, the authors show that indeed SVEP1 binds to PEAR1 with high affinity, and this affinity drops my many folds in the presence of PEAR1 missense variant that associate with SVEP1 in trans. Then, the authors go on to map the molecular pathways downstream of SVEP1-PEAR1 binding: AKT and mTOR signaling in vascular smooth muscle cells.
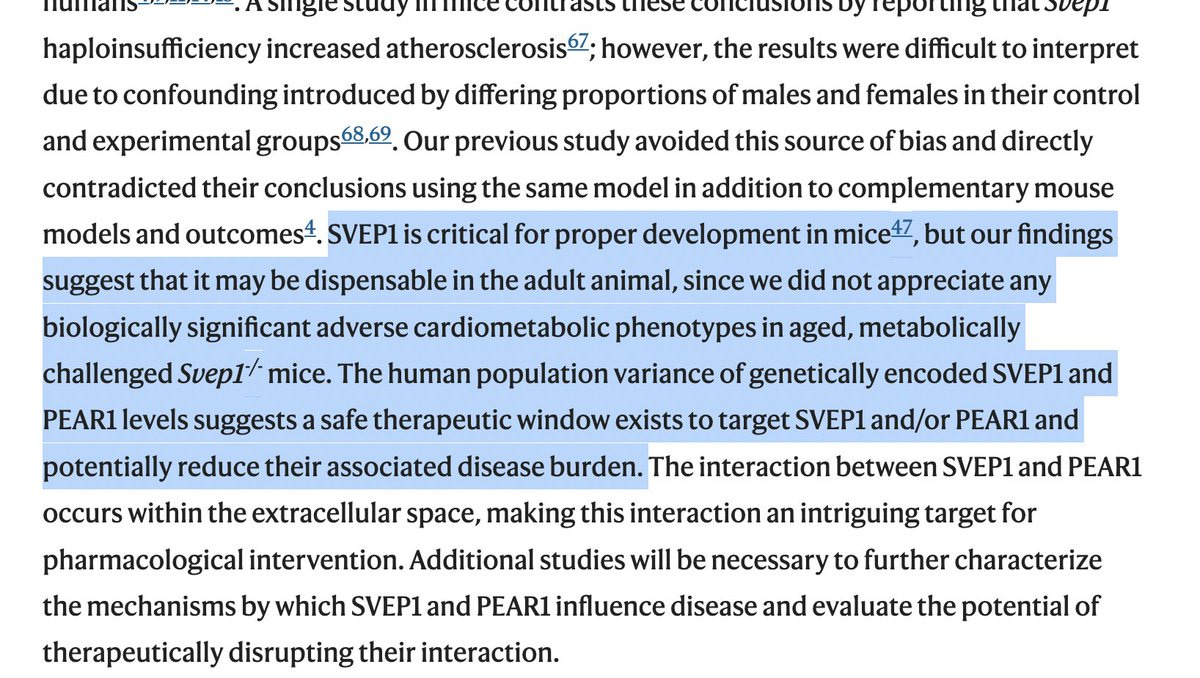
Given that human genetic studies suggest that SVEP1 inhibition might be cardioprotective, the authors studied SVEP1 KO mice. While loss of SVEP1 is lethal, partial loss seems to be well tolerated in mice in normal and disease backgrounds. But there were no signs of cardioprotection.
Given the strong human genetic associations that increased SVEP1 is deleterious, the authors suspect that "a safe therapeutic window exists to target SVEP1 and/or PEAR1 and potentially reduce their associated disease"
Though therapeutic value of the findings is still not clear, the realization of SVEP1 as the physiological ligand for PEAR1 was extremely fascinating. A great example of human genetic findings leading to incredible biological insight.