Genetics of British Pakistanis
Genomes of Brits of Pakistani descent opens a window into the life, culture and health history of South Asians
Happy Friday! I’ve been thinking about underrepresentation of South Asian population in growing genomic databases. Although the situation is still bad—South Asians continue to be underrepresented in genome-wide association studies—it is better than before. There are many locally-led efforts such as Pakistani Genomics Resource, GenomeAsia 100k consortium etc. that are stretching their arms to fill the big diversity gap. But there are also many efforts in the UK and US, where researchers are building South Asian genetic cohorts by recruiting Brits and Americans of South Asian origin, respectively. One such cohort is Genes and Health, a mini UK Biobank of South Asians in the UK, which has been recruiting British-Pakistanis and British-Bangladeshis for several years now with the goal of reaching 100,000 participants in the next few years. For this week’s From the Twitter archives post, I highlight a Twitter thread I wrote two years ago on one of the many interesting works that came out of the Genes and Health study.
From the Twitter archives
This is an important work from Elena Arciero, Hilary Martin and colleagues from Wellcome Sanger Institute, UK on the fine-scale population structure and demographic history of British Pakistanis
I am glad I stumbled upon this while I am in India visiting family after a long time and, as usual, contemplating about my culture and the impact it had on my genes.
South-Asian populations have rich and complicated cultural histories, traces of which can be found in today's south-Asian genomes (something that scares the South-Asian me, but fascinates the geneticist me).
Here, the authors studied the genetic structure of 2,200 British Pakistanis of South Asian ancestry, "one of the largest and most socioeconomically disadvantaged ethnic minorities in the UK".
Like Indians, Pakistanis also identify themselves in distinct caste communities, which determine their social status in the community. People marry only within their community and this endogamy is being practiced for hundreds of years, and it is believed to have become stronger during the British rule.
These hundreds of years of endogamous practices have populated South-Asians’ genomes with numerous recessive mutations, making them the world's best population to mine for human knockouts.
Endogamy can be genetically quantified using IBD scores (number of segments of genome that are identical across two or more individuals, hence come from a common recent ancestor), which will be higher in endogamous communities.
The authors show that the IBD scores of British Pakistani communities are many orders of magnitude higher compare to Finns, a well studied founder population who underwent multiple bottleneck events.
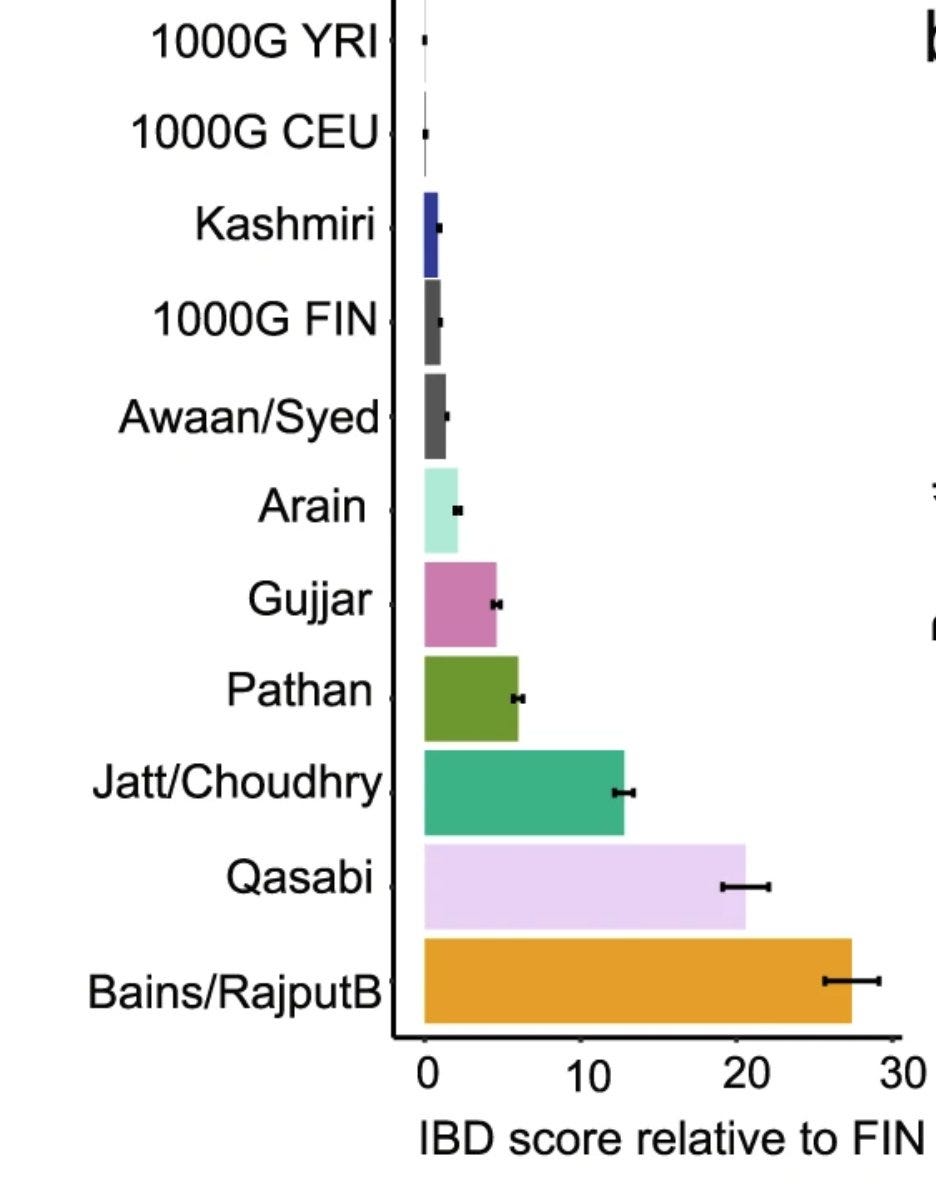
A similar plot showing IBD scores of Indian caste communities relative to Finns and Ashkenazi Jews reported by Nakatsuka et al. in 2017 in Nature Genetics, a landmark paper in the area of Indian population genetics.

One finding that I found particularly fascinating is the difference in the ROH (regions of homozygosity) between those with both parents born in Pakistan vs those with one parent born in Pakistan and the other in US.
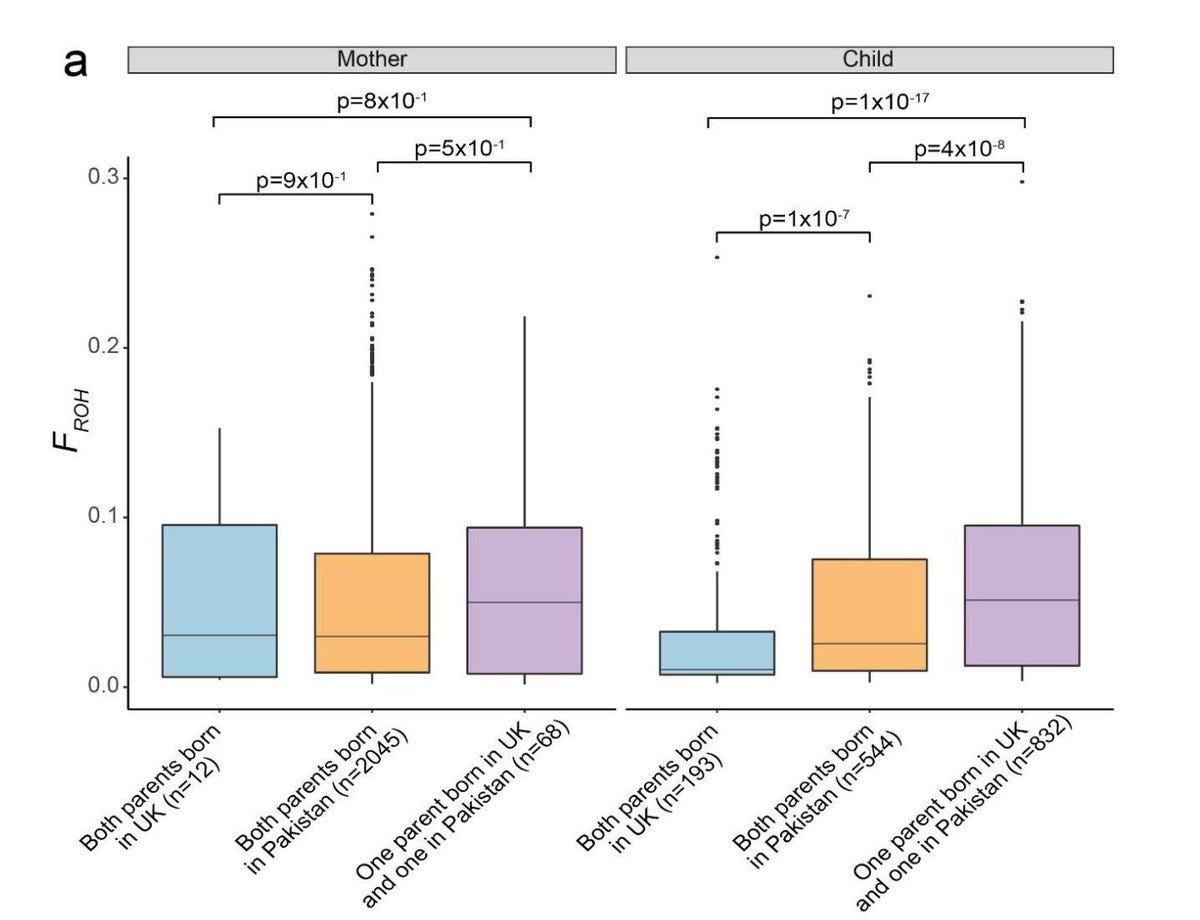
A higher ROH for one parent born in UK and one in Pakistan compared to both parents born in Pakistan should come as a surprise to many. But this is likely due to that British Pakistani marriages often transnational. In a review of the anthropological literature on Pakistani migration and settlement published in Human Heredity in 2014, the author Alison Shaw from University of Oxford writes
British Pakistani marriages often involve a partner from Pakistan who joins a spouse in the UK. Transnational marriage of first cousins offers relatives in Pakistan opportunities for a 'better' life in the West and are important for British Pakistanis for economic, social, cultural and emotional reasons."
This is a fascinating example of how cultural practices can impact our genomes, and a reminder that such factors are important when studying genetics of non-European ancestries.
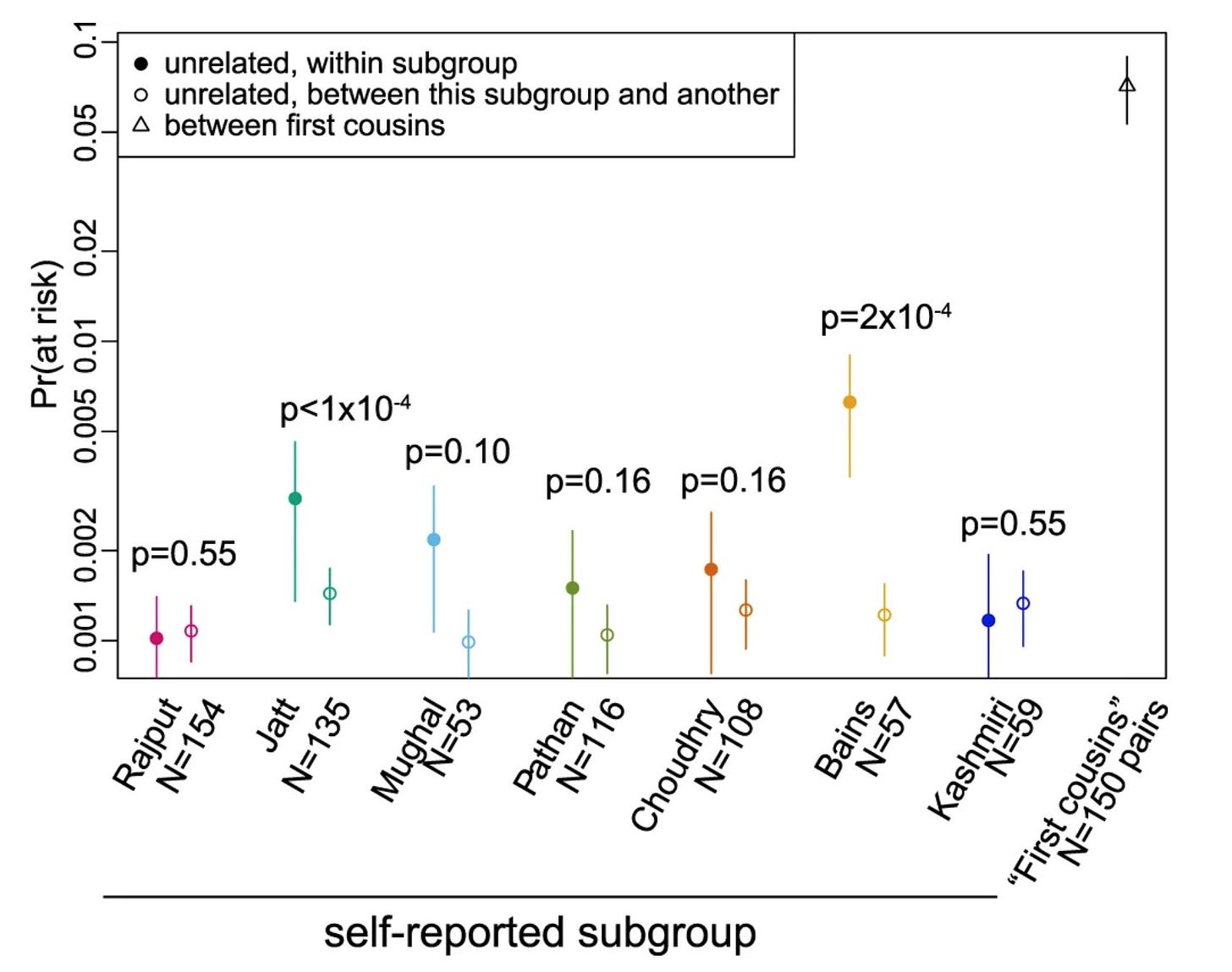
Then, using info about known pathogenic variants identified from the exome data of a subset of these participants, the authors simulate endogamous and consanguineous unions and predict the risk for autosomal recessive conditions in the offspring using an approach from a recent paper in AJHG by Fridman et al.
The simulations show that the risk for giving rise to an offspring with recessive genetic conditions are higher for within community marriages compared to between community marriages, particularly in the communities with high IBD scores.
Putting these results in context, one could imagine how highly prevalent must be these recessive diseases in many of the South-Asian communities, where endogamous and consanguineous marriages continue even today. I couldn't resist imagining those ceremonious marriages ending in a lifelong suffering for the couples who will spend the rest of their life traveling to clinics, chasing doctors with the hope that one day they'll find a cure for their child's condition.
That is one side of the story, the sad side, particularly for me as a South-Asian who is born and brought up in one such community. But there is another side to this story, the side that excites and fascinates the geneticist in me who is in constant search for those rare gifted--or often cursed--humans, who might be holding the secret to treat/cure human diseases.
I've tweeted about many of those rare humans who were identified among the South-Asian communities.
The first example that comes to my mind is a young boy from Pakistan, born in a consanguineous family, who was found to be a human knockout for gene SCN9A and helped scientist discover the role of this gene in sensing pain. Sadly, he met with a tragic end. Believing he was invincible, the boy jumped off a house roof and lost his life. The story is told beautifully in the book Genome Odyssey by Euan Ashley.
The second example is a healthy adult British-Pakistani woman, a participant of the Genes and Health Cohort in the UK, who was found to be a knockout for a gene HAO1, the RNAi target of Lumasiran developed by Alnylam for the treatment of primary hyperoxaluria type 1. Studying this woman’s health profile, drug developers ensured that complete inhibition of this gene is safe. In 2020, the drug became the first ever FDA-approved RNAi medicine for a rare disease.
The third example is a recent discovery of the association of rare loss of function variants in MC3R with age at menarche. A major highlight of the study is the identification of a human knockout for MC3R, a British Bangladeshi, from the Genes and Health cohort who helped scientists learn about the critical role of MC3R in human growth and puberty.
Many more such fascinating stories exist showing that people of South-Asian communities, due to their unique cultural history, are one of the world's best populations to do human genetics. I hope more South-Asian populations are represented in the future genetic studies, and importantly I wish such studies benefit those communities by creating awareness of the harmful endogamous practices and improving genetic diagnosis and counseling.
It's Sunday morning. I just had my first filter coffee. I still haven't recovered from my jet lag.
As I turn through the pages of today's Hindu paper, I couldn't resist stopping at the matrimonial section to share a few snapshots. I couldn't have wished for better images to relate to my thread.







According to Razib Khan, endogomy is relatively rare in Bangladesh unlike the rest of South Asia. Most Bangladeshis have similar genetics, as in there is a lot of cross community marriages in the region.
Wonderful read as always! Rare diseases unfortunately remain neglected in India and so does the scientific know how of how to avoid them. Wrote a piece on it a while back and thought of sharing it here: https://genomeofindia.substack.com/p/genome-10-udgam-of-india-a-genetic