Null mutations in interferon genes in Polynesia
Discovery of the founder mutations responsible for vaccination- and viral infection-associated deaths in Western Polynesians
Happy Friday! The aftereffects of the controversial UMAP figure in the All of Us flagship publication in Nature last week are still lingering on Twitter. The reactions on Twitter to a scientific figure (that was created by the authors with good intentions to showcase the rich diversity of the All of Us biobank) have made me worry about how the woke mentality of certain scientists might hurt the very communities that these people are trying to help. After years of outcry on the unfairness of genome [white] association studies, we finally have a remarkably diverse biobank which is expected to fuel genetic discoveries for the next decade or so that will benefit many communities who were historically underrepresented in human genetic studies. What was supposed to be a celebration turned into a PR nightmare for AllofUS after a famous geneticist posted a comment on Twitter that UMAP was a bad choice to represent the diversity of All of Us and might be misinterpreted by the public. To be honest, it was a fair criticism. However, the extent to which the scientific community reacted by anchoring on that comment and expressing their hatred towards the authors and the human genetics field, in general, was unfair.
Two days ago, a new wave of Tweets and threads erupted after some celebrity population geneticists in the field for some reason decided to express their pedantic opinion about the phrase “ancestry-specific variant” used in human genetics studies. They argue that, in the human populations, no variants can be truly ancestry-specific, and if you search enough you can find all variants in all populations; it is not the case that there are many variants that we will miss if we do not sequence every ancestry in the world. As a person from an underrepresented population, who has been voicing the importance of diversity in human genetic studies for a long time, I couldn’t resist expressing my thoughts on Twitter. While the comments may be fair in their truest sense and I am sure they were made with the best intentions in mind, I am worried about the potential of such comments to encourage those who hate genetics to undermine the importance of studying diverse populations.
So, given these happenings, I thought this Tweet thread that I wrote two years ago on the discovery of certain loss of function mutations in the genes part of the critical immune response pathway that are enriched in individuals of Western Polynesian ancestry is a good choice for this week’s From the Twitter archives post. I like this story because it not only highlights the medical importance of discovering ancestry-enriched variants but also highlights the importance of such discoveries in impacting policy changes at the national level.
From the Twitter archives
Genetic discoveries such as this are mind-blowing not only because they are fascinating--both clinically and biologically--but they also demand immediate attention from policymakers.

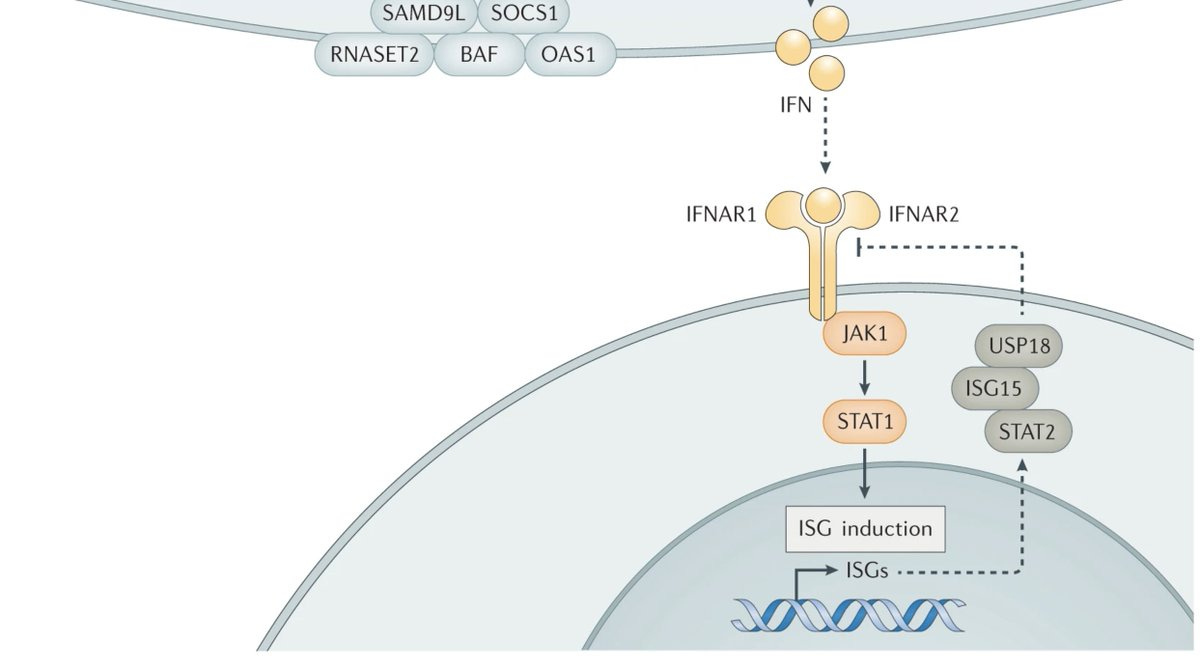
Two papers published side by side in the Journal of Experimental Medicine report rare deleterious mutations in IFNAR1 and IFNAR2 that abolish the type 1 interferon signalling, the pathway critical for mounting an anti-viral response upon natural viral infection or vaccination.
In the first paper, Bastard et al report a case series of seven children of West Polynesian ancestry who are all knockouts for IFNAR1 (that codes for a subunit of type 1 IFN receptor) due to a same homozygous nonsense mutation.
This variant is almost non-existent in most parts of the world but has an MAF of 0.025 in Western Polynesia (which would translate into roughly 0.5-1% of homozygotes who are human knockouts for IFNAR1). Though it's tempting to assume that there might be a beneficial effect that resulted in positive selection, the authors write the high MAF is likely due to genetic drift.
Loss of IFNAR1 in these 7 patients resulted in extreme susceptibility to viral infections, even the less virulent forms such as the one in live attenuated vaccines (LAV). The case report of the seven children described in the paper is heartbreaking to read.
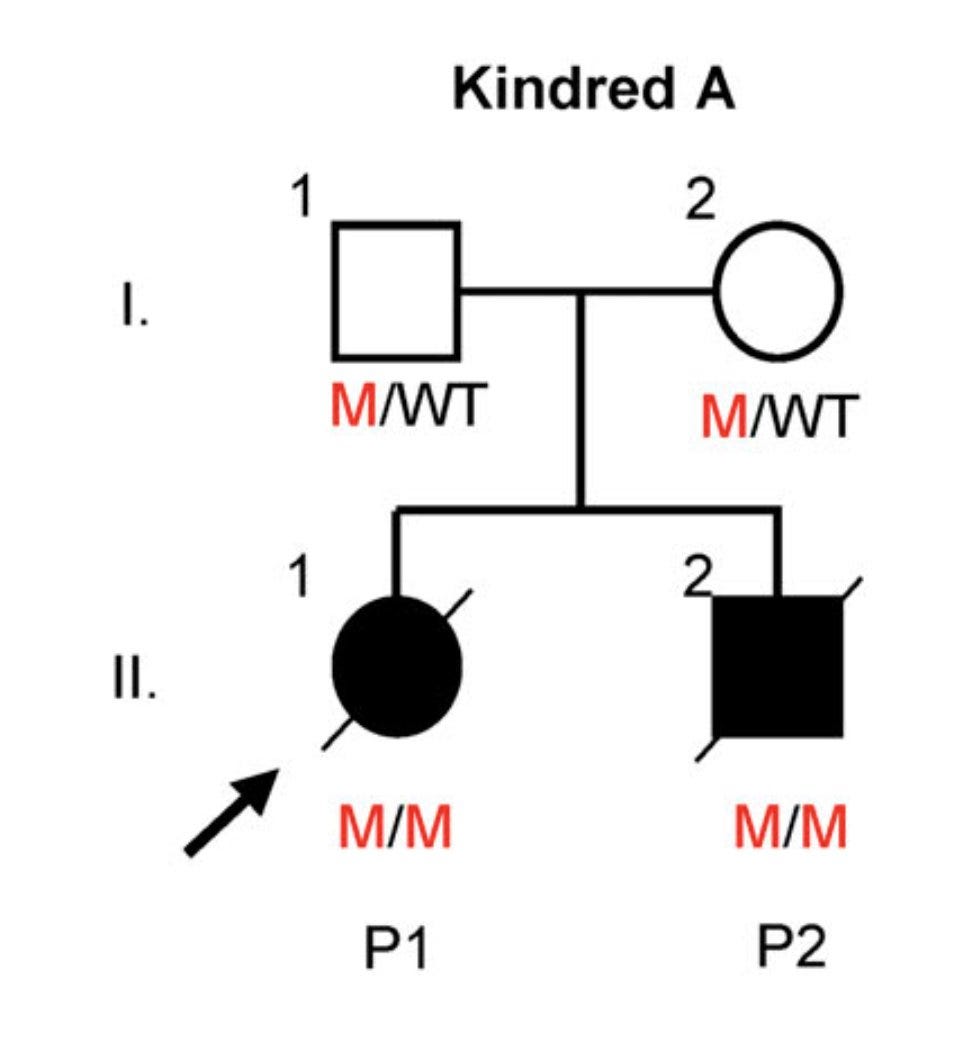
Patient 1 (P1) was a 1-year-old girl who developed a severe injection site reaction on day 5 after MMR vaccination, deteriorated over the next few days and died on day 18 with multi-organ failure.
P2 is the older brother of P1 who also met with the same fate--died on day 21 after the first MMR vaccine. This family is so unlucky because the chance of both the children inheriting a disease allele from heterozygous parents is 1 in 16 (1/4*1/4).
P3 is a 15-month-old girl who developed encephalopathy after the MMR vaccine, continued to deteriorate despite aggressive treatment and died on day 72. Notably, the child had a history of enteroviral meningoencephalitis and mild ID at 2 months of age.
P4 is a 13-month-old boy, a preterm child, who developed epilepsy, encephalopathy and ARDS on day 14 after the first MMR vaccine. The boy fought and recovered after 4 months of hospitalization only to die later of a respiratory syncytial virus infection.
P4 has a sibling P5 who also fell ill 17 d after MMR but recovered. Then again got meningitis from enterovirus at 2yrs from which too he recovered. Surprisingly he tolerated 2nd dose of MMR. He's now 7 years old with no neurological deficits except for the hearing loss due to mumps.
P6 and P7 who are currently alive also had a similar clinical history as others involving multiple episodes of severe viral infections and prolonged hospitalizations.
The nonsense mutation (p.Glu386*) results in protein truncation and loss of cell surface expression. Skin cells of one of the pts confirmed the loss of type 1 interferon exposure. Thereby the authors confirm that all 7 children have autosomal recessive complete IFNAR1 deficiency.
In the second paper, Duncan et al report a case series of 5 children of Inuit ancestry all with the same homozygous missense mutation in IFNAR2 (that codes for the other subunit of type 1 IFN receptor) resulting in complete loss of IFNAR2 function.
The five children with IFNAR2 mutation had strikingly similar clinical histories as the 7 children with IFNAR1 mutation reported in the earlier paper. In addition, two of the children also had severe COVID-19 infection, of which one died.
Like before, this mutation also results in complete loss of gene function—loss of cell surface expression and loss of response to IFN-1. In addition, the authors also show that the defects can be rescued by introducing the wild-type allele.
Interestingly, this variant seems to be present at high frequency (MAF-0.024) in the circumpolar region (Greenland, Alaska, Nunavik) whose present-day inhabitants descend from common ancestors who migrated to this region ~1000 years ago, hence suggestive of a founder effect.
So, the two papers combined report 12 cases with either IFNAR1 or IFNAR2 deficiency all from geographically isolated populations. It's now extremely important that these populations are studied at scale and required policy changes are made.
Both papers clearly state that these findings by no means undermine the importance of MMR and other vaccinations as the mortality due to not vaccinating will be undoubtedly higher than the mortality due to vaccinations. However, national screening programs must be set in place in these populations to identify those with IFNAR genetic deficiencies and provide alternative methods of protection such as monoclonal antibodies, mRNA vaccines or novel antiviral agents.