Some interesting papers that I read recently
Let's talk about IGF1R, JAK2, IgA and Africa
I am still figuring out how to structure the posts for GWAS stories. For now, I am not sticking to any particular structure. I’ll let the posts evolve and shape on their own over time. For today, I’ll share some interesting findings that I came across recently.
IGF1R, a new type 2 diabetes risk gene identified based on the exome sequencing data from the UK Biobank
An interesting convergence of genetic associations with aortic disease between germline non-coding variants and a somatic coding variant in JAK2
Ugandan genomic resource, an African cohort of 5000 individuals that is increasing the representation of African ancestries in human genetic studies
A fascinating GWAS of serum IgA levels which for the first time finds that serum IgA levels are higher in African ancestries compared to other ancestries—both phenotypically and genetically.
IGF1R, a novel type 2 diabetes risk gene
UK Biobank is a gift that keeps on giving. Since the release of UK Biobank exome sequencing data, we have now many iterations of rare variant association analysis of type 2 diabetes (T2D). The latest is from Eugene et al published in Cell Genomics. This is a great paper, and it has many fascinating findings. The major finding is the uncovering of a beautiful association between rare deleterious missense variants in IGF1R and T2D. It’s interesting that others who did T2D exome-wide association study (ExWAS) previously in the UK Biobank have missed it. According to the authors, this finding became prominent when testing burden associations with only highly deleterious variants.
The authors report that individuals who carry rare deleterious missense variants in IGF1R (n carriers=394), particularly variants within a specific protein domain (protein tyrosine kinase domain), have 2.4 folds increased odds of T2D. IGF1R codes for the insulin-like growth factor 1 receptor to which the IGF1 binds and exerts its actions. There is fascinating biology underneath this finding.
It turns out these individuals who carry defective IGF1R proteins have elevated levels of circulating IGF1 levels, which is a feedback consequence (as the circulating IGF1s are not doing their job, the system assumes that there is an IGF1 deficiency and starts producing more IGF1). Elevated levels of IGF1 would mean that the carriers are taller than the non-carriers as IGF1 mediates the action of growth hormone, and that is what we see in individuals with acromegaly. But it turns out the carriers are on average 2.2 cm shorter than the non-carriers despite having elevated levels of IGF1, suggesting that these individuals have IGF1 resistance.
The molecular mechanism of how the missense variants disrupt the IGF1R function is even more fascinating. Unlike missense variants, the heterozygous loss of function variants in IGF1R have no effect on T2D risk or on height. The authors speculate that this is because, in the case of missense variants, the defective transcripts get translated into defective proteins that can still participate in dimerization leading to defective receptor complexes. On the other hand, in case of loss of function variants, the defective transcripts are probably getting eliminated by non-sense mediated decay. Though no experiments have been done to prove this, the hypothesis makes sense as we have seen similar mechanisms in other genetic conditions, for example, PCDH19 disorder, an X-linked disease in which only partial gene loss is lethal but not complete gene loss. As a result, only heterozygous females are affected, but homozygous females and hemizygous males are spared.
One last finding that I want to highlight is the challenge in performing Mendelian Randomization analysis using genetic variants associated with phenotypes such as IGF1 that are under feedback regulation. If we do a GWAS for IGF1 and identify genetic variants that are associated with increased levels of IGF1, we should be aware that these variants can represent at least two distinct sets: one that associates with increased levels of biologically active IGF1 and one that associates with increased levels of biologically inactive IGF1 (i.e., with IGF1 resistance). These two groups of variants can have completely opposite effects on downstream phenotypes and cancel out each other. The authors illustrate this by identifying two IGF1-increasing common variants—one near the IGF1 locus (rs11111274) and the other near the IGF1R locus (rs1815009). These two variants seem to have directionally opposite effects on childhood height and T2D risk. Hence, the authors conclude, “reported common variant instruments for higher IGF-1 levels comprise a mixture of functionally opposing signals, i.e., higher levels of bioactive IGF-1 or higher IGF-1 resistance.”
A convergence of genetic effects between germline and somatic variants
One way to identify the causal gene in a GWAS locus is to study the phenotypic consequence of activating or inactivating the genes in the vicinity of the GWAS locus in humans. Large-scale ExWAS enable us to do this. I often highlight the PIEZO1 GWAS locus associated with varicose veins as an example. One of the beautiful findings that emerged from UK Biobank exome data was the association of heterozygous loss of function variants in PIEZO1 with increased risk for varicose veins and also, an association of what appears to be a gain-of-function missense variant in PIEZO1 with decreased risk for varicose veins. So, by studying the phenotype of individuals with defective and overactive PIEZO1 channels, it became clear that PIEZO1 was indeed the causal gene at the GWAS locus associated with varicose veins.
So, to do this sort of investigation, it’s clear that we need to gather many carriers of functional coding variants. Apart from sequencing a large number of individuals, what else would help us to identify carriers of functional coding variants? We can go study special populations where some functional coding variants rise to high frequency due to population phenomena such as genetic drift or natural selection. A recent paper made me realize that such special populations perhaps might also include cancer patients in whom certain somatic mutations rise to high frequency.
An interesting paper published in Nature Communications reports a detailed investigation of the association of a common gain-of-function somatic mutation (V617F) in JAK2 with aortic aneurysm. JAK2 codes for a cell signaling protein that is essential for many cell surface receptors to function properly. JAK2 V617F is the most common somatic mutation in JAK2, and it causes the over-activation of receptors involved in red blood cell and platelet differentiation from progenitor cells in bone marrow leading to hematologic malignancies. Particularly the overproduction of red blood cells (the condition described as polycythemia vera) is due to the over-activation erythropoietin receptor (encoded by EPOR) in bone marrow cells by the hormone erythropoietin produced in the kidney. It seems the JAK2 mutation also has an interesting extrahematopoietic consequence: aortic dilatation and aneurysm, which are commonly seen in individuals with the JAK2 mutation. Through elegant experiments, here the authors show that this effect is mainly due to the over-activation of JAK2 signaling in aortic tissue-resident macrophages that also express EPOR and are targets of circulating erythropoietin. The authors also demonstrate that over-infusion of erythropoietin alone is sufficient to cause aortic dilatation without the JAK2 mutation, a finding that has been also shown previously, with the motivation coming from a clinical case of aortic aneurysm with a history of chronic hemodialysis and repeated injections of recombinant erythropoietin.
Reading the paper, I noticed the authors mentioned a supporting germline association between JAK2 intronic variants and aortic aneurysm reported by a GWAS. So, the current study is confirming that the causal gene at the GWAS locus for aortic aneurysm is indeed JAK2. Although this GWAS finding is not why they did this study in the first place, I led the Twitter thread with the GWAS finding just to highlight how the somatic variant association in this case has brought insight into the GWAS association. I wonder how often we might be able to capture convergence in the genetic effects between somatic coding and germline non-coding variants. I guess it’s an unexplored territory and has the potential to offer valuable insights into many of the GWAS findings.
Ugandan genomic resource
One of the highlights of this year’s American Society of human genetics (ASHG) conference is the presidential symposium on African genomics. An excellent line of speakers gave inspiring presentations on the latest advances in human genetics happening on the African continent. Large-scale biobanks are being built. Hundreds of African scientists are getting trained in genomic analysis. Fantastic registers are being established for some of the common diseases in Africa such as sickle cell disease (SCD) to enable participation in clinical trials at a continent-wide scale. Francis Collins gave an update on an ambitious initiative that NIH launched in 2019 in collaboration with the Gates Foundation to cure HIV and SCD in Africa through in vivo gene therapies (note, existing treatments involve removing patients’ blood cells, gene editing them, and putting them back inside, which requires a high-quality healthcare setup, something that’s not available in under-developed countries). The project is progressing faster than I expected, said Francis Collins.
With these highlights still fresh in my memory, I came across a paper published in Cell Genomics describing an African cohort for genomic studies (BTW some high-quality content is coming out in Cell Genomics. Kudos to the editors). Reading the article, I realized that I have tweeted about a couple of studies based on this cohort, but never took the effort to read about the source of the study samples. It was refreshing to read about study participants for once rather than the genetic findings, and it was inspiring to learn how such invaluable resources are built in underdeveloped countries with minimal manpower and funding support. Such cohorts can be easily overlooked in today’s world of big biobanks. But it’s extremely important, particularly for young researchers, to understand and appreciate the efforts happening in countries like Africa.
The paper describes an African genomic database called “Ugandan genomic resource” that has today DNA array based genotype data for ~5000 individuals and whole genome sequencing data for ~2000 individuals. This cohort was built from a base cohort called Ugandan general population cohort (GPC) which was founded in 1989 by the Medical Research Council, UK in collaboration with the Uganda Virus Research Institute. The GPC was originally established to study the epidemiology of HIV infection, a fact that stood out when I was scanning through the cohort characteristics: 7.6% of the participants in the UGR are HIV positive (a number many folds higher compared to western countries). It reminded me of how disproportionately African continent has been hit by the HIV epidemic.
I also enjoyed reading the community engagement section. It seems the GPC has a community advisory group comprised of members who are the leaders of the community. They have built dedicated office spaces for the sole purpose of community engagement activities like dissemination of research findings, discussion about new study procedures etc.
The UGR participants were genotyped using 2.5M Illumina chip array yielding around ~2.2 million good quality variants for imputations. They even have built their own imputation reference panel based on their in-house WGS data (n=2000) combined with 1000 genomes WGS data (n=2504). They have identified 41.5 million variants based on their WGS data and around 9.5 million of them are never seen before, highlighting the incredible diversity of the African genomes. The resource has contributed to many GWAS already and importantly, the summary statistics of such studies were deposited in the GWAS catalog for anyone to access freely, a gesture that deserves appreciation.
Genetic differences in IgA between African and European ancestries
Speaking of African ancestries, I read an interesting paper reporting a GWAS of serum levels of IgA, an immunoglobulin subtype that plays an important role in mucosal immunity. The highlight of the paper is the discovery of increased IgA levels in individuals of African ancestries compared to other ancestries. Looking at the genetics of IgA, the authors speculate that the increased IgA levels in African ancestries could be at least partly due to an enrichment of IgA increasing genetic variants in Africans compared to Europeans (or alternatively, due to a depletion of IgA increasing genetic variants in Europeans compared to Africans; it could be either a positive selection in Africans or a negative selection in Europeans).
The phenotypic differences are compelling. The authors demonstrate significantly high IgA levels in African ancestries compared to Europeans in two independent cohorts. Further, they also demonstrate a moderate correlation between African ancestry proportion and serum IgA levels in an admixed African American cohort (this analysis is less likely to be affected by environmental confounders and highlights the value of admixed populations in human genetic studies).
The authors performed a multi-ancestry GWAS of IgA in ~22k individuals and further meta-analyzed the results with an independent cohort of ~19k individuals from a previous study for which they had access to only a subset of genetic variants (the authors of the previous study seem to have shared publicly only significant and suggestive associations). In the final meta-analysis, the authors identify 20 genome-wide significant loci of which 11 are novel.
The beauty of doing GWAS for a biomarker phenotype such as serum IgA levels is that the causal genes and the mechanisms are often easily predictable in most of the loci. For example, the strongest association was seen at a locus in chromosome 1 near gene RUNX3; the index variant is located upstream of RUNX3, and the minor allele reduces IgA levels by 0.80 SD, the largest effect size observed in the study. The relevance of RUNX3 to serum IgA levels can be readily appreciated if you read this abstract from an in vitro IgA study.
What is even more compelling is that the GWAS captured signals not just for RUNX3, but also for its interacting partner RUNX2. Both the transcription factors appear to be essential for IgA antibody class switching.
Just next to RUNX2 locus in chromosome 6 is a locus near gene CITED2, which encodes a protein that acts as a molecular switch in the TGF-α and TGF-β signaling. If you go back to the abstract above again, you’ll realize that TGF-β induces IgA production. It’s all connected, and the GWAS of IgA is enabling us to reconstruct parts of those molecular connections. That’s the power of GWAS that many failing to appreciate. And that’s why I always love GWAS of molecular traits such as blood biomarkers like IgA.
Okay, let’s come back to the major finding: ancestral difference in serum IgA levels. Motivated by the compelling phenotypic difference in the IgA levels between African and European ancestries, the authors explored the differences in the allele frequencies of IgA associated variants across ancestries. Impressively, at 12 (60%) out of 20 GWAS loci, the IgA increasing alleles are more frequent in African than European ancestries. The most striking difference was seen in a locus near GPATCH2, which showed the second largest effect size next to RUNX3 and surfaced only because the authors studied African ancestries. The index variant at this locus is seen in ~10% of Africans but almost absent (~0.02%) in Europeans.
Not only the IgA increasing variants are enriched in Africans, it also seems the IgA decreasing variants are depleted in Africans, for example, the IgA decreasing RUNX3 variant is common in Europeans (MAF ~1.2%) but rare in Africans (~0.2%).
Wait, you haven’t heard the most interesting part of this story yet.
So, what is the clinical relevance of IgA antibody? There are so many, given the IgA’s role in immunity. But there is one particular condition that immediately comes to mind when a clinician hears the word IgA—IgA nephropathy, an interesting auto-immune condition characterized by production of antibody against antibody—IgA and IgG auto-antibodies are produced against the IgA antibody. The resulting antibody-antibody complexes deposits in the glomerulus of the kidneys causing chronic kidney disease and renal failure.
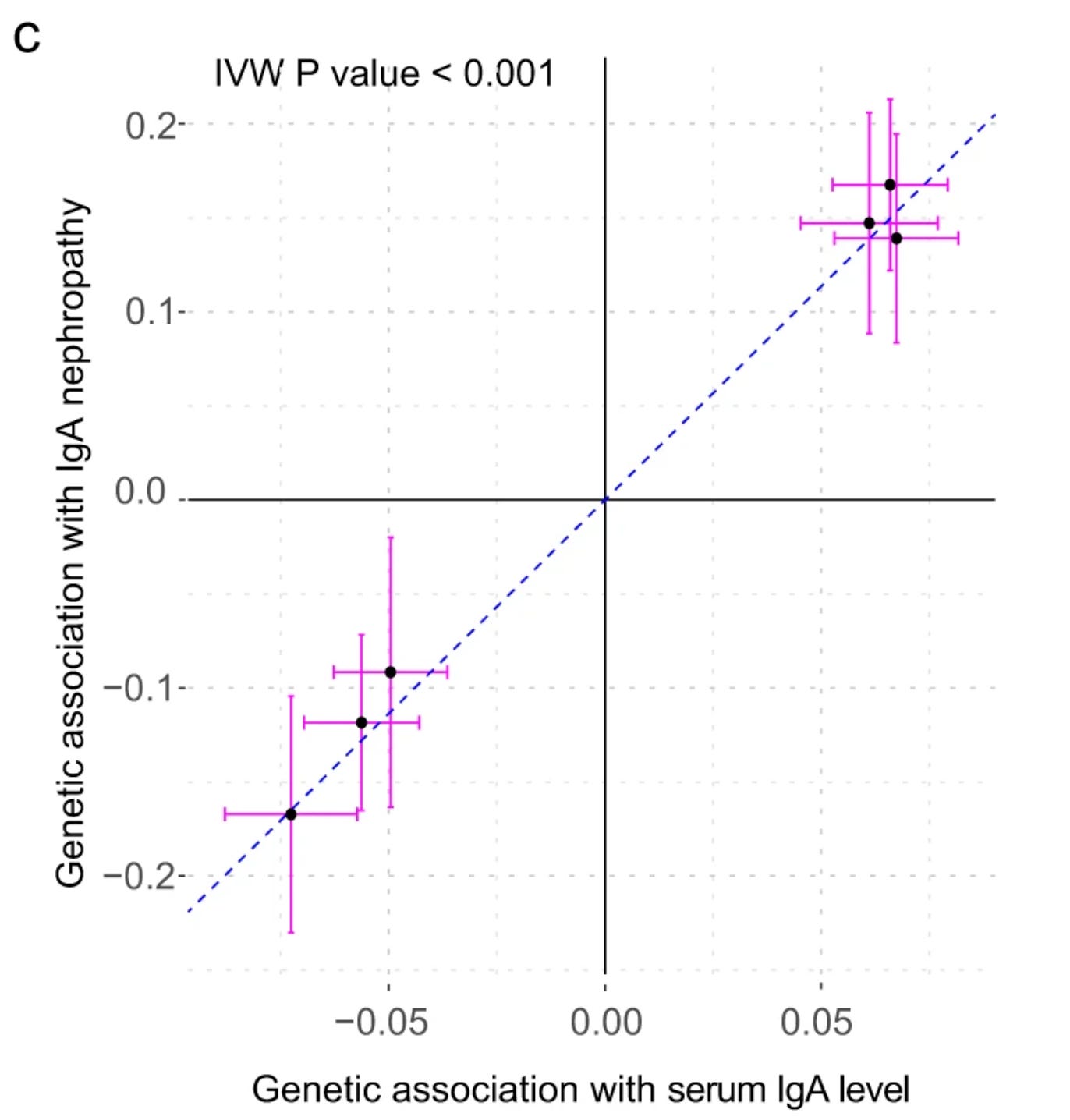
Although the pathogenesis of IgA nephropathy is not clearly understood, increased production of IgA antibodies lies in the causal path to kidney disease. And those genetically predisposed to produce more IgA antibodies are at higher risk for IgA nephropathy, the causality of which can be tested now as we have many strong, mechanistically clear GWAS loci for serum IgA levels. Through Mendelian Randomization, the authors indeed show a strong causal relationship between serum IgA levels and IgA nephropathy.
Given the above background, you’d expect that IgA nephropathy is more common in African than European ancestries. It’s not. IgA nephropathy is rarely seen in individuals of African or African American ancestries. The authors also have a parallel manuscript on GWAS of IgA nephropathy (which is still in preprint) in which they write,
“Notably, IgAN is less frequent among individuals of African ancestry, including African Americans, suggesting that protective genetic effects may exist, but further studies are needed to address this hypothesis.”
This discrepancy makes this finding even more interesting. One thing to note is that in the IgA nephropathy the autoantibodies are targeted towards a specific subtype of IgA that have lost a carbohydrate tag that says to the immune system ‘I’m not foreign’. It’s not clear at the moment why individuals of African ancestries, despite having a high IgA blood level, are rarely found with IgA nephropathy. There’s definitely more to this story, and we’ll learn about it eventually.
That’s all for now. I am working on a long post (which I’ll probably break into two or three parts) based on my recent talk on the rare variants and drug target discovery. I might be able to post the part 1 in a week or so.
Happy Thanksgiving :)
—Veera