The cost of waking a gene up from its eternal sleep
Noncoding de novo mutations release beta cell-specific repression of hexokinase to cause congenital hyperinsulinism
Happy Friday! Before we jump on to this week’s story, I’d like to share something. Some of you might have heard the 2022 and 2023 year-end episodes of The Genetics Podcast, hosted by Patrick Short, the co-founder and CEO of Sano Genetics, where I talked about the most exciting genetics papers of the respective years. Motivated by the positive feedbacks, the Sano Genetics team has asked me to do quarterly roundups this year. The first one was published yesterday, in which I talk about some of the stories that I’ve written about in my earlier Substack posts this year. Here is the link, if any of you would like to hear it. The main theme of the Q1 roundup is ‘noncoding variants’, one of my favorite topics. One of the flavors of papers that I excitedly watch out for is noncoding discoveries and their underlying mechanisms. There is a whole magical biological universe buried in our noncoding genomes, and I can’t wait for rapidly growing whole genome sequencing efforts (both short and long reads) to uncover that universe layer by layer. Every noncoding discovery has an underlying, unique, beautiful story that needs to be told to the world, like the one below, which I wrote in Twitter two years ago.
From the Twitter archives
Genetic discoveries like this derepress my hidden desire to become a clinical geneticist :) A beautiful revelation of a set of non-coding mutations that allow a disallowed gene to express in beta cells leading to congenital hyperinsulinism.
There are so many reasons why I absolutely love this paper. The finding takes me on a trip down my memory lane to the days when I taught biochemistry to medical students.
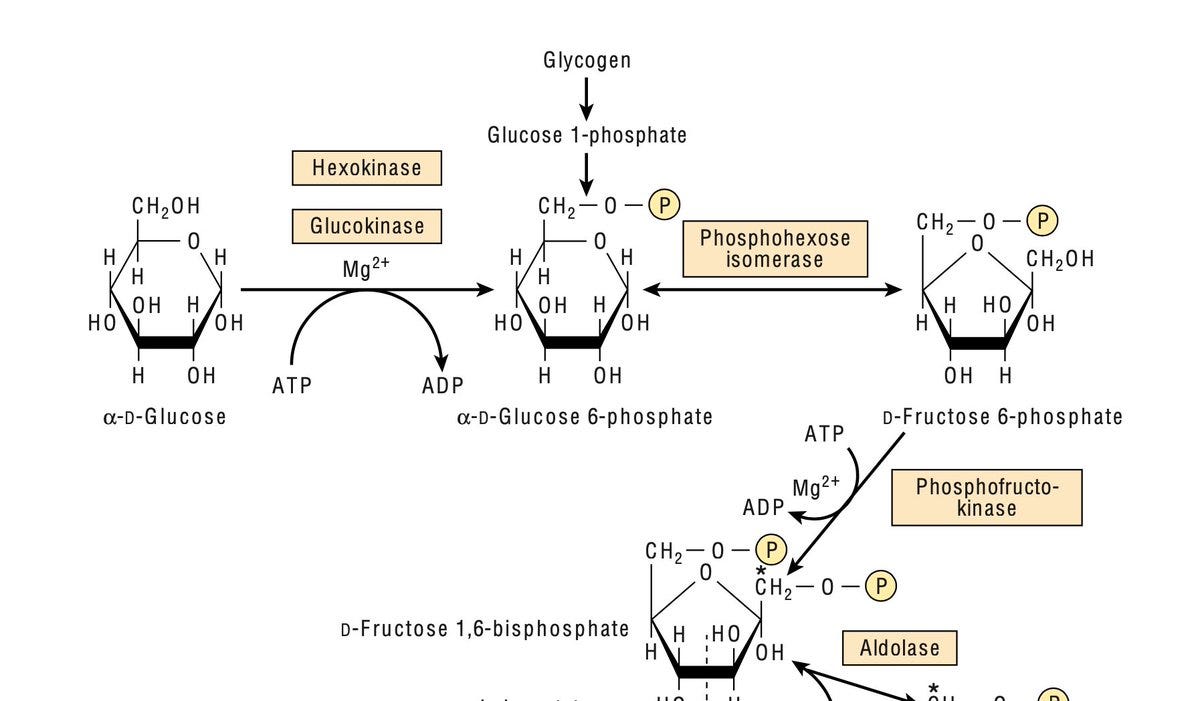
The first step of glycolysis, the key pathway of glucose metabolism in almost every human cell, is the conversion of glucose to glucose-6-phosphate catalyzed by hexokinase.
Hexokinase type I (HK1) is ubiquitously expressed throughout the body, except liver and pancreas, where instead of hexokinase type I, we have hexokinase type IV, commonly known as glucokinase (notice the left-most two tissues in the below plot from GTEx)

Nature has assigned glucokinase instead of hexokinase in beta cells for a reason. Hexokinase's high affinity for glucose (low Km) makes it fully saturated at all the time, thereby ensuring constant supply of energy to body cells. Whereas, glucokinase's low affinity for glucose (high Km) in beta cells ensures insulin is secreted only when glucose concentration is higher than normal.

Students confuse the two often. We'd ask the student to imagine the real world consequence of their mistake that is reversing the roles of glucokinase and hexokinase.
Actually, you don’t have to imagine, natural experiments via human genetic variations have already illustrated this in real humans. Rare gain of function mutations in GCK abnormally increases glucokinase's affinity for glucose, making it behave like hexokinase, leading to congenital hyperinsulinism (Osbak et al. Human Mutat 2009)
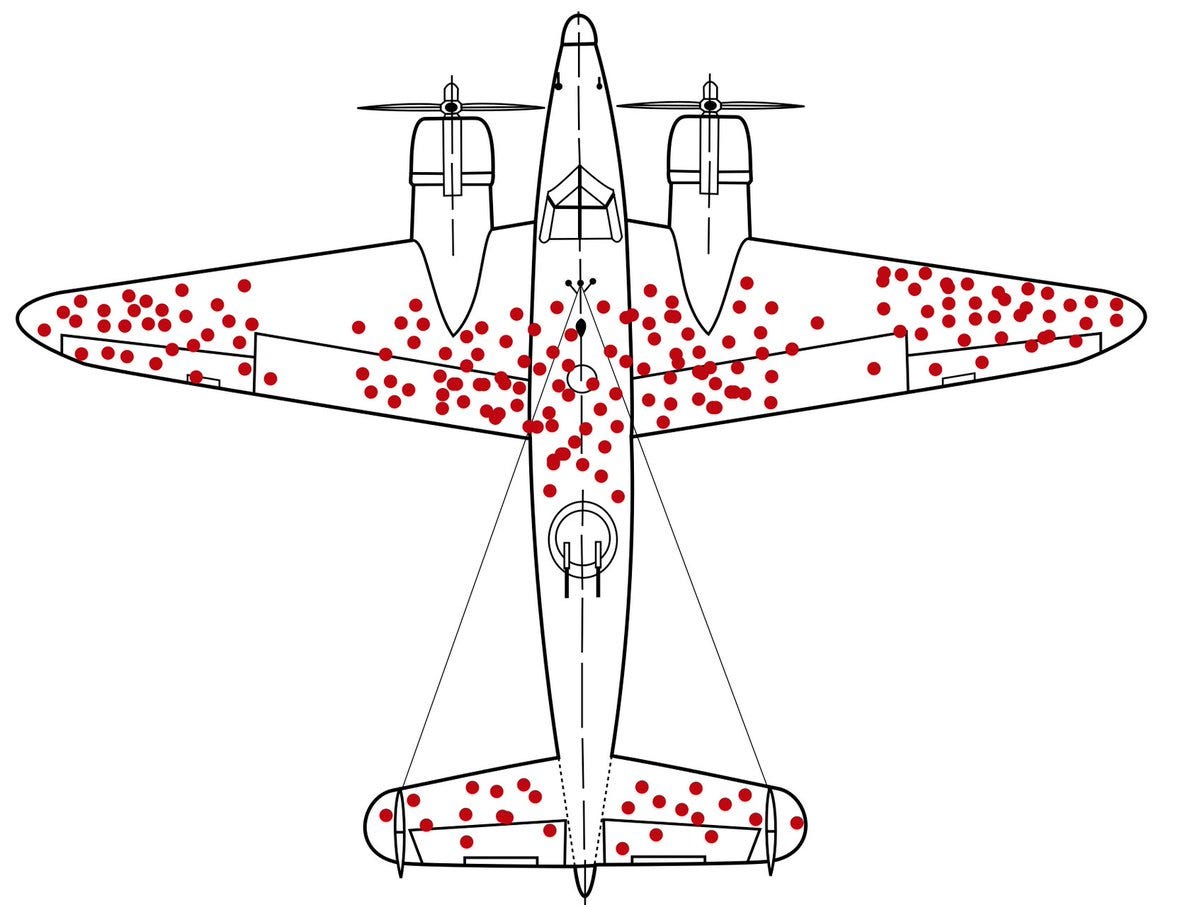
In the current paper, for the first time, the authors discovered not just one but 7 different denovo mutations and a ~4.5kb deletion CNV within intron 2 of hexokinase causing congenital hyperinsulinism. It turns out that there is a 42bp region within intron 2 of HK1 that is highly constrained, meaning, it is depleted of mutations like the unhit regions of this plane.
Transcription factors persistently bind to this region in beta cells and liver cells and repress HK1 expression. Mutations hitting this critical region awakens HK1 from its eternal sleep in beta cells. As a result, hexokinase takes back control of glycolysis from glucokinase in beta cells, leading to persistent glycolysis and hence uncontrolled chaotic insulin secretion.
What a beautiful finding! Even more interesting is coding mutations in HK1 affects RBCs, but not beta cells as the gene is never expressed in beta cells in normal conditions, which makes HK1 a unique case where coding and non-coding mutations in a gene causing distinct phenotypes in distinct cell types.
When we talk about non-coding variants, we always talk about tissue specific expression, but rarely about tissue specific repression. The current discovery points out the survivorship bias in our thinking. Sometimes the answer might lie not within tissues where a gene is expressed, but within tissues where a gene is not expressed.