A gene x gene interaction study of alcohol behavior in East Asians
Regions in the human genome that interact with rs671, a missense variant in ALDH2, and influence alcohol behavior and cancer risk in East Asians.
Happy Weekend! Here is an interesting GWAS story to enhance your weekend science reading.
Gene-environment (GxE) and gene-gene (GxG) interactions have always been of great interest to human geneticists for ages, even before genome-wide association studies (GWAS) were a thing. One can pull hundreds of hypothesis-based GxG and GxE studies from the literature, most would be from before the GWAS era when scientists studied one gene at a time based on prior hypotheses. But none of them survived the test of time (except a few, of course, like the ALDH2 variant, which is the topic of this post1). In hindsight, one could see why: lack of statistical power.
To identify GxE and GxG associations for a variant, two things are important.
High allele frequency. The variant is sufficiently common so that when you stratify based on genotypes, you get enough sample size in each group (at least in wild types and heterozygotes)
Large effect size. The variant has a large effect size which makes the characteristics of the genotype subgroups sufficiently different to identify any genetic or environmental differences between them.
There are only very few variants in the genome known to date that are common (either globally or in specific populations) and have a large effect size (they are often described as "low-hanging fruits"), most of which were identified during the early GWAS era. A couple of examples: a missense variant (I148M) in PNPLA3 associated with fatty liver and a missense variant rs6712 in ALDH2 associated with alcohol consumption in East Asians.
I’ve written Twitter threads in the past about the discovery of PNPLA3's association with fatty liver. It’s one of my special interests as it is actively being pursued as a drug target by many companies, including Alnylam Pharmaceuticals in collaboration with Regeneron3. Last year, I also wrote on Twitter about a study that uncovered an interesting GxSex interaction showing that the PNPLA3 I148M variant damages the liver more severely in women compared to men. PNPLA3 is one of the topics that I might be writing a deep dive post in the GWAS Stories in future.
Coming back to our current topic, a new study has just dropped in Science Advances which reports a comprehensive investigation of genome-wide GxG interactions for the famous East-Asian specific ALDH2 variant rs671.
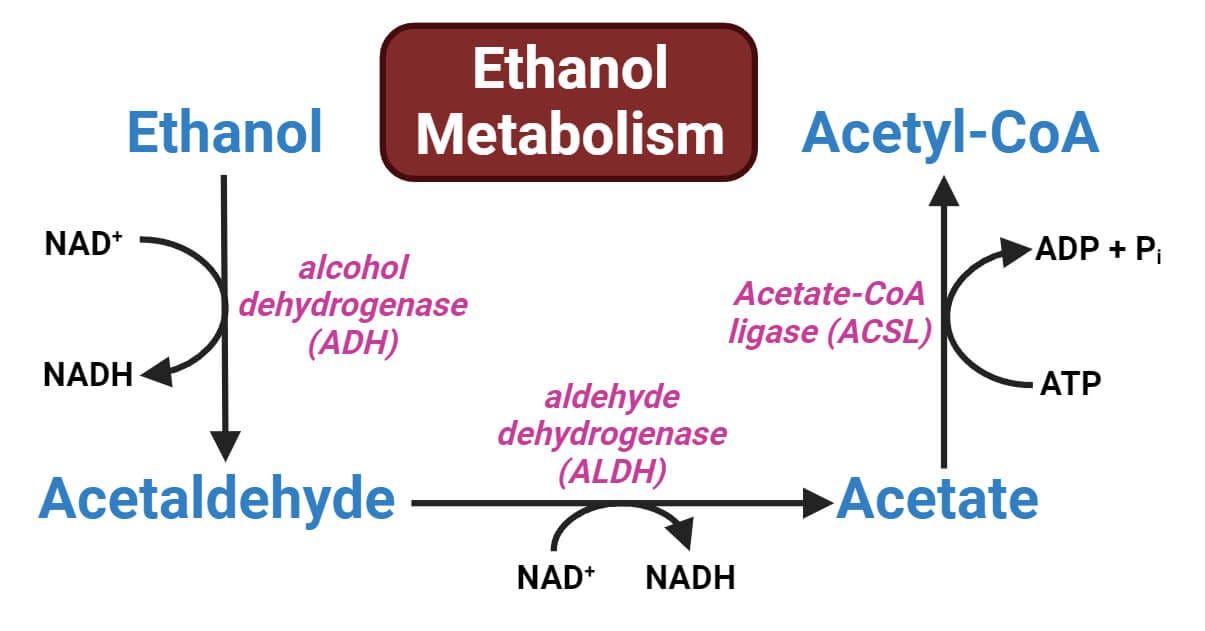
ALDH2 encodes acetaldehyde dehydrogenase that detoxifies acetaldehyde, an intermediate product of alcohol metabolism. The SNP rs671 almost fully inactivates the enzyme thereby rendering its carriers (mostly East Asians as the variant is absent in other populations) intolerant to alcohol.
Apart from influencing alcohol behaviour, rs671 is associated with a wide range of diseases and traits, most of which are believed to be downstream effects of alcohol intake.4 One such association is the risk of oesophageal cancer which is seen almost exclusively in heterozygotes who manage to drink alcohol tolerating the acetaldehyde. The increased levels of acetaldehyde (which is a carcinogen, by the way) put them at higher risk of getting cancer. This is one of the most beautiful GxE interactions known in the human genetics literature.5
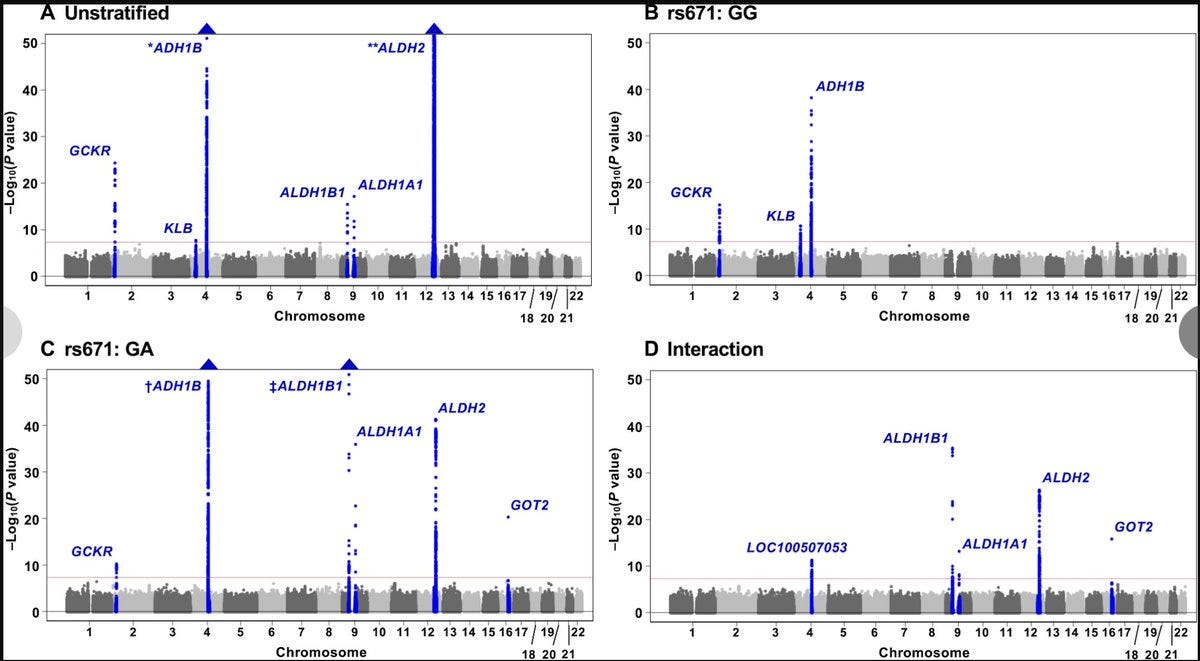
In the current study, the authors explored GxG and GxGxE interactions of rs671 in Japanese individuals. They stratified the study individuals into wild type (GG) and heterozygotes (GA) and performed GWAS of alcohol intake in each group separately (AA homozygotes are relatively few and among them, alcohol drinkers are very few due to extreme unpleasantness, hence not suitable for a stratified analysis).
The stratified GWAS identified many interesting genotype-specific associations including one right at the rs671 locus itself (which the authors argue is driven by a variant that is in complex LD with rs671 but tags an independent risk haplotype6). The authors then show that these GWAS signals show significant GxG interaction with rs671.
Among those that interact with rs671 is a variant in ADH1B encoding another enzyme alcohol dehydrogenase that catalyzes the conversion of alcohol to acetaldehyde. ADH1B locus is a well-known alcohol locus in both East Asians and Europeans (unlike the ALDH2 variant, the ADH1B variant occurs globally, though with wildly different allele frequencies7).
Though ADH1B x ALDH2 interaction has been long hypothesized (given the perfect biological rationale), this is the first time someone has empirically demonstrated the interaction effect (you can't do this in European populations).
The authors further studied associations with oesophageal cancer risk in individual genotype groups and found many known alcohol GWAS signals showing an association with cancer risk in heterozygotes but not in wild types, confirming the fact that alcohol drinking increases cancer risk only in heterozygotes.
One other finding that I find striking is the difference in the genetic correlation for alcohol behaviour between Europeans and East Asians when stratified by genotypes. While there is a high genetic correlation (97%) between East-Asians rs671 wild types and Europeans, the genetic correlation drops abruptly between East-Asians rs671 heterozygotes and Europeans (28%), which makes sense8, but fascinating to see in real data.
Overall, this is a beautiful example of a genetic study with tons of insights that can never be found by studying Europeans even in a million individuals, highlighting the importance of studying non-European populations.
After I published this post on Twitter, I received a comment from Prof. George Davey Smith (a pioneer of the Mendelian Randomization method) who pointed out that some parts of the post are misleading and shared articles from the past that have reported GxE interactions for rs671. I didn’t intend to mislead the reader into thinking this is the first GxG or GxE interaction study of rs671. What I wanted to say was that the human genetics literature from the pre and early GWAS era is littered with many hypothesis-driven GxG and GxE studies most of which never turned out to be true. But, of course, there are few well-designed candidate gene studies with strong biological rationale like the ones George pointed out.
The ALDH2 missense variant rs671’s association with alcohol intake was known long before the GWAS era and is the model of the mechanism of action of disulfiram used in the treatment of alcohol use disorder.
If you’d like to learn more, tune into episode 5 of The Genetics Podcast where Josh Friedman, a research scientist from Alnylam talks about Alnylam’s PNPLA3 RNAi program.
But recently functional studies have been surfacing showing that rs671 might have alcohol-independent effects, for example, a mouse knockout study that I summarized last year on Twitter showed that knocking in the rs671 variant in mice resulted in an increased risk for diet-induced obesity contrasting the human genetics finding that rs671 protects against obesity (likely due to low alcohol consumption).
Another beautiful example is the CHRNA5 x Smoking interaction influencing the risk of lung cancer, a topic that is near and dear to Kari Stefansson from deCODE genetics, a GWAS pioneer and one of the first to map the first GWAS locus for smoking and lung cancer.
I must admit that I haven’t fully understood the discussion of this rs671 cis GxG interaction. It looks like an independent haplotype but I am not sure.
The minor allele of rs1229884 in AHD1B in East Asians is the major allele in Europeans and vice versa.
Makes sense in the way that the alcohol behaviour of rs671 heterozygotes East Asians will be strikingly different from that of Europeans (who are all wild types) which will result in a different genetic architecture of alcohol intake.