Discovery of a new obesity gene--Bassoon
Analyzing more than half a million exomes, scientists find that haploinsufficiency of a less known synaptic gene, BSN, causes extreme obesity
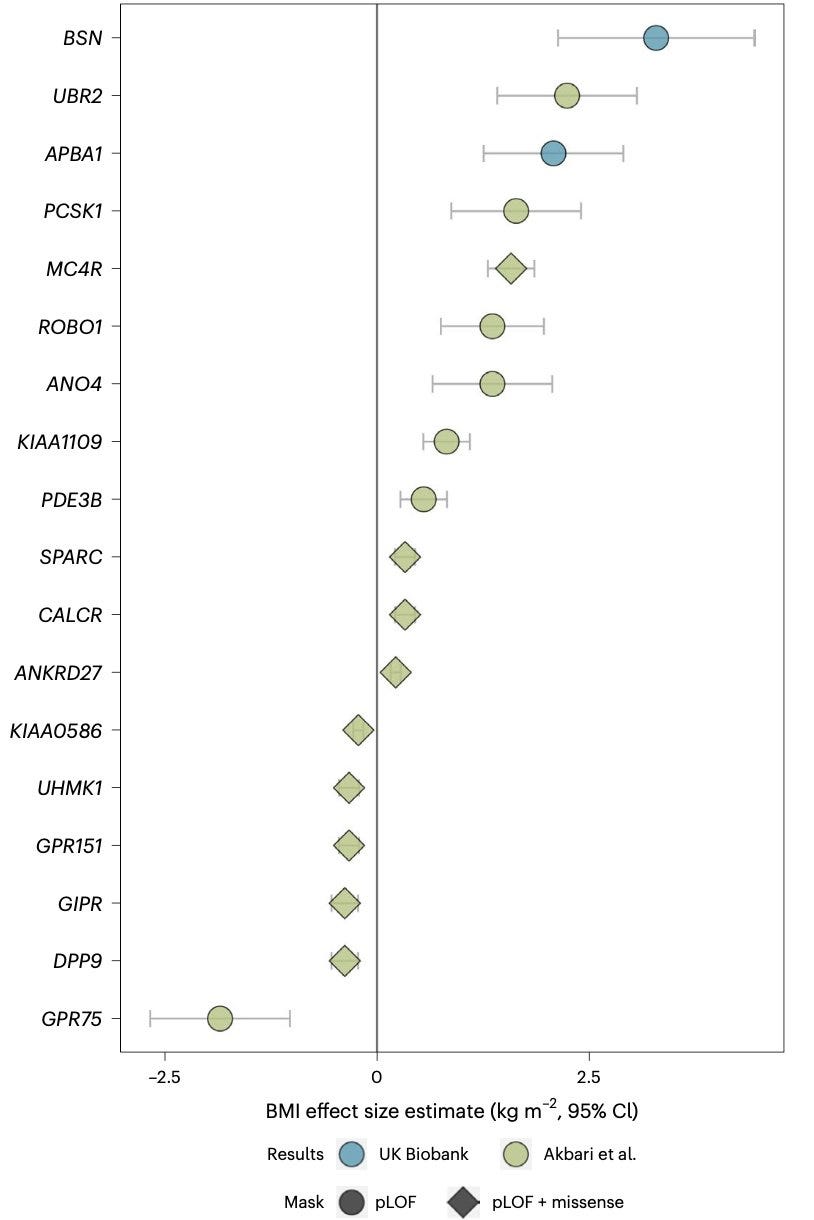
Happy weekend! Here is a bonus weekend read for you. In a new paper, published in Nature Genetics, scientists from MRC Epidemiology Unit, University of Cambridge, report a new obesity gene—BSN. Predicted loss of function (pLOF) mutations in BSN are linked to an extreme form of obesity that is reminiscent of the weight gain linked to mutations in Mendelian genes like LEP, LEPR, POMC, MC4R etc. The authors find that individuals with haploinsufficiency of BSN are even more obese than those with haploinsufficiency of dominant obesity genes like MC4R.
This is not the first exome-wide association analysis (ExWAS) of BMI in the UK Biobank. An early ExWAS of BMI published in Science by scientists from Regeneron in a much larger sample apparently missed reporting this gene. It is not surprising, given that there are some uncertainties in the way we aggregate rare variants across a gene to study their collective impact on a phenotype. It seems that Zhao et al.'s bucket of BSN pLOF carriers had 65 individuals, yielding a P value of 2e-8 whereas Akbari et al.'s bucket of BSN pLOF carriers had slightly different numbers of carriers (I am guessing more), yielding a P value that was less significant.
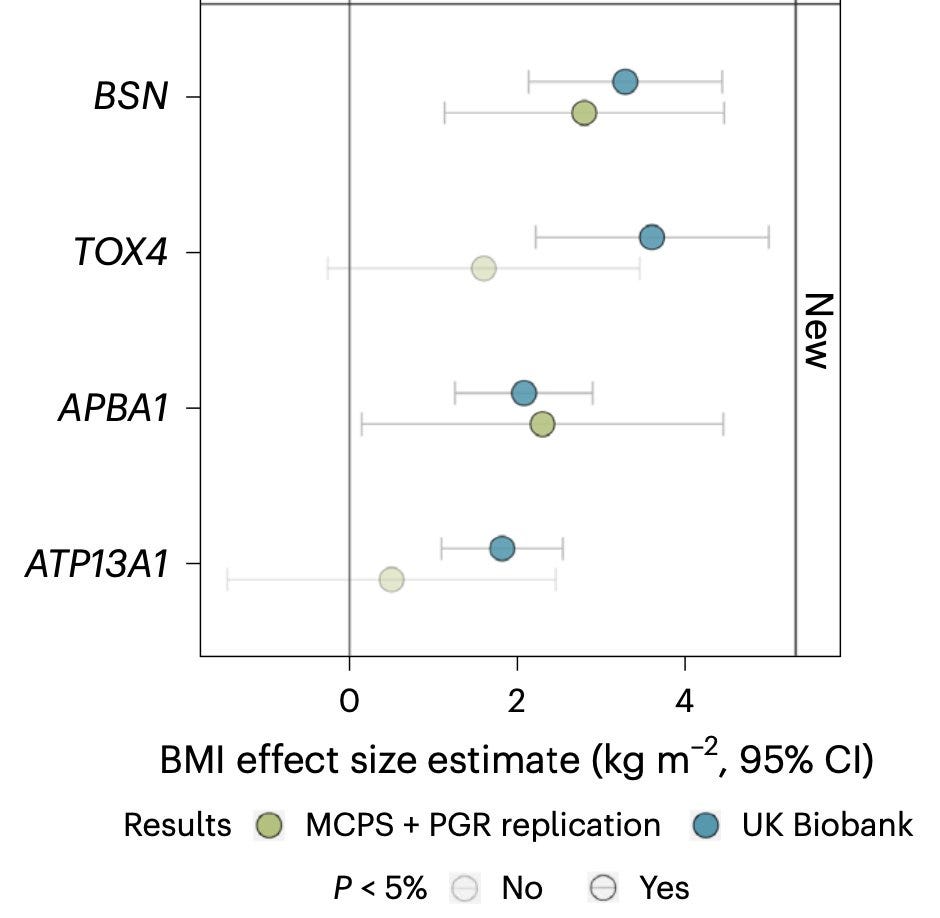
You'll appreciate the inherent instability of P values from rare variant aggregate analysis, when you look at an earlier report in NPJ Genomic Medicine by scientists from the Columbia University, who also independently identified the association of BSN with obesity in the UK Biobank in a relatively smaller sample size. Zhu et al. analyzed only 1/3 of the UK Biobank cohort and so, should have found a lot fewer carriers than Zhao et al. or Akbari et al., yet they landed in a much stronger P value of 3.6e-12. It's likely that their bucket contained individuals carrying more severe, highly penetrant, BSN pLOFs than the other two buckets, perhaps simply by luck. It is clear that BSN association with obesity is a real one as it beautifully replicates in independent cohorts (Mexican Americans from MCPS and Pakistanis from PGR cohorts), but it's interesting that how variant filtering criteria influences the gene discovery.
While many in the field are wondering why the initial analysis of UK Biobank exomes did not find BSN, I am pondering over the question of why we did not know long ago about this gene, which seems to have a profound influence on BMI. It's really an interesting thing to think about.
The BSN discovery is made through a "hypothesis-free" approach, that is, you are searching through the entire genome (exome, in this case) blindfold and discovering genes whose biological links to obesity we know nothing about. After the discovery, we try to make sense of how loss of this gene cause obesity, slowly realizing the role of new biological pathway(s) involved in obesity.
This is in striking contrast to the classic hypothesis-driven approach. Some of the greatest discoveries in the field of obesity genetics (made by some of the senior scientists from the current paper) were made in the 1990s via hypothesis-driven approach. Mice genetics were instrumental behind those hypotheses that resulted in the world of knowledge of obesity biology that we consume today. And you'll be pleasantly surprised to find that how much serendipity played a role in the early mouse genetics discoveries that later inspired human genetics discoveries.
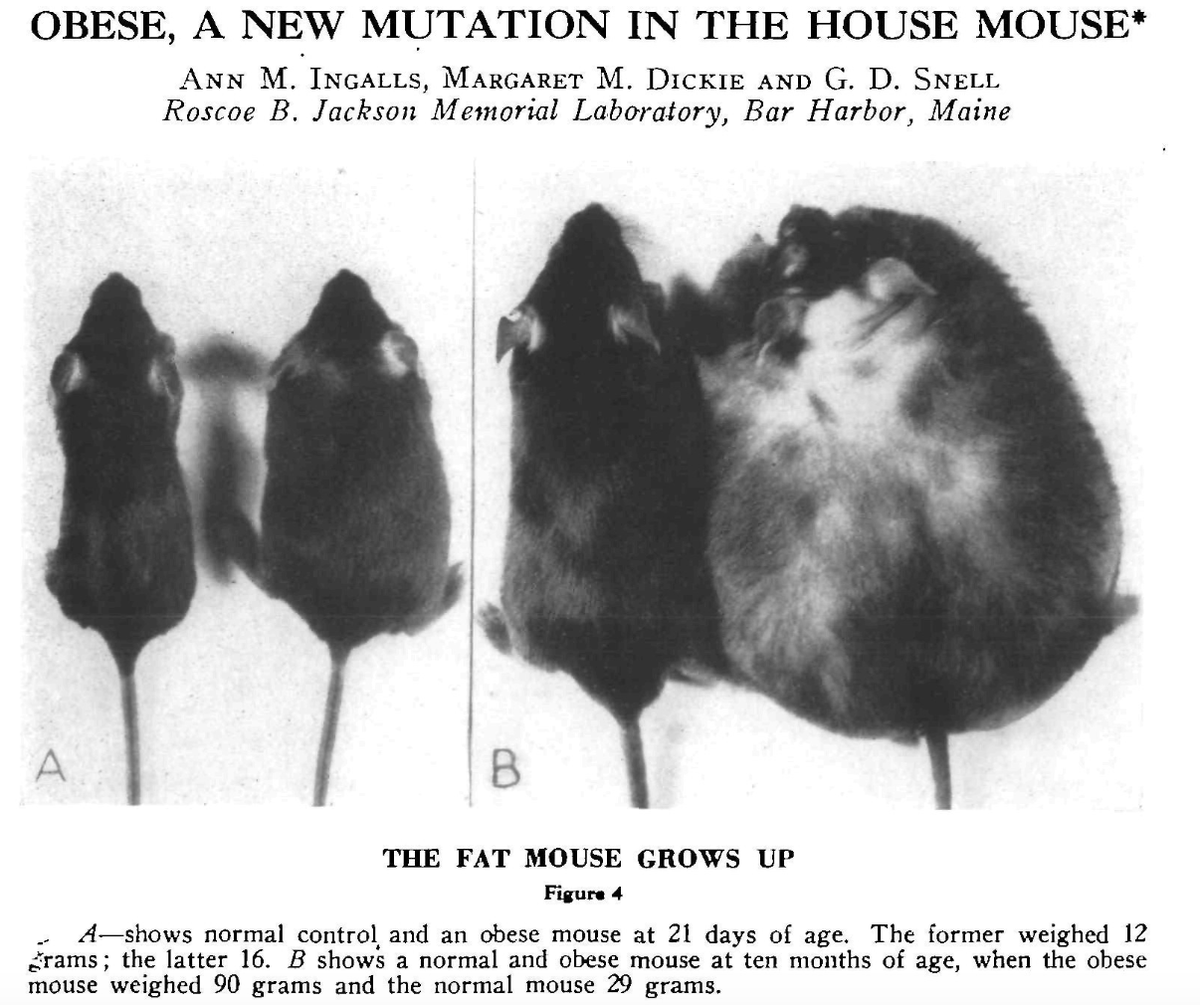
"In the summer of 1949 some very plump young mice were found in V stock" reads a 1950 paper in the Journal of Heredity published by Jackson Laboratory scientists. That was the first report of a naturally occurring obesity-causing mutation in mice, and appropriately named "ob" (Agouti yellow obese mice were known even before that, probably in the early 1900s, but the spontaneous mutation underlying Agouti obese mice was not found until the 1990s).
Later in the 1990s, genetic advancements led to the cloning of ob gene in mice and its homologue in human leading to the discovery of leptin hormone secreted by the adipose tissue and eventually, to the discovery of leptin receptor in brain hypothalamic neurons that regulate the appetite.
Once the biology was out in the open, it was just a matter of time before scientists got their hands on the first human counterparts of ob/ob and db/db mice (caused by leptin receptor mutation). All they had to was keep their eyes open and be ready to act immediately when the opportunity presents itself in the form of obese humans seeking medical attention.
Fate would have it, the first fruit of obesity genes in humans fell in mid 1990s on the hands of Stephen O'Rahilly, a physician scientist at the University of Cambridge, shaping his influential career in the endocrinology and metabolism for the next decades. Studying two severely obese children from a consanguineous family, Stephen and his team discovered the very first genetic cause of obesity in humans--congenital leptin deficiency--toppling the first domino of many that followed immediately in the late 1990s.
Discovery of first leptin receptor homozygous mutation in humans by French scientists in 1998 by sequencing the LEPR gene in a morbidly obese child from a consanguineous family.
Discovery of the first POMC homozygous mutation by German scientists in 1998 by sequencing two patients with obesity, adrenal insufficiency and red hair pigmentation.
Discovery of the first MC4R mutations by two independent teams—one from University of Cambridge and the other from Institut Pasteur de Lille, Paris.
The contrast between the MC4R discovery and the recent discoveries of GPR75 and BSN is what inspired me to sit and type this post. Yeo, Farooqi et al. studied 63 severely obese children and specifically looked for mutations in MC4R that might explain their high BMI, finding a heterozygous frameshift mutation in one. Vaisse et al. studied 43 morbidly obese individuals and specifically looked for mutations in MC4R and found a heterozygous frameshift mutation in one. In contrast, in the hypothesis-free approach, Zhao et al., after scanning the whole exomes of more than half a million individuals, captured 65 carriers of heterozygous loss of function mutations in BSN with extreme obesity. I think if not for the understanding of leptin and melanocortin pathways in appetite regulation, it would have taken many more years for scientists to discover LEP, LEPR, POMC and MC4R mutations in humans via a hypothesis-free manner.
The current BSN discovery probably opens a new biological pathway that plays an important role in human appetite regulation, a pathway that is also linked to neurodegeneration. BSN encodes a neuronal scaffolding protein critical for presynaptic cytoskeletal organization. The gene was first cloned in 1998 almost at the same time as other classic obesity genes. However, it was discovered in the context of neurodegeneration. Studying the gene expressions in the brains of patients with multiple system atrophy, Japanese scientists stumbled upon a novel transcript leading to the cloning of BSN gene (which was called ZNF231 initially). Should this discovery had happened based on the brains of obese humans, BSN's link with obesity would have been discovered in the 1990s.
It's fascinating to think how profoundly influential were prior knowledge and hypotheses on the timeline of human genetic discoveries.