Insights from a deep dive into the genetics of Lp(a)
Genetic variants in LPA teach us about missing heritability, genetic differences between ancestries and allelic series
Happy Friday! For this week’s From the Twitter archives, I picked a thread I wrote on the beautiful genetics of Lp(a) based on a paper by Ronen E. Mukamel, Po-Ru Loh, and colleagues from Harvard Medical School published in Science in 2021. This is one of the two papers that discovered a variable number tandem repeat (VNTR) in ACAN associated with height and the prequel to Po-Ru Loh’s team’s work on long-read sequencing-based VNTR imputation in the UK Biobank published in Cell last year, which I have discussed in my previous posts, including the recent Feb roundup.
From the Twitter archives
A lovely paper that I enjoyed reading again in its final form. (no surprise Science published it along with a great commentary).
Let me take this opportunity to revisit the wonderful genetic insights that we can extract from a single gene—LPA.
LPA codes for lipoprotein(a) (Lp(a)), one of the very few proteins in humans whose expression levels are almost completely genetically determined. More than half of the Lp(a) variation is due to a variable number tandem repeat (VNTR). Information about this VNTR for almost half a million individuals along with data on common and rare variants in the right hands can lead to extraordinary insights—a realization that will strike anyone who reads this paper.
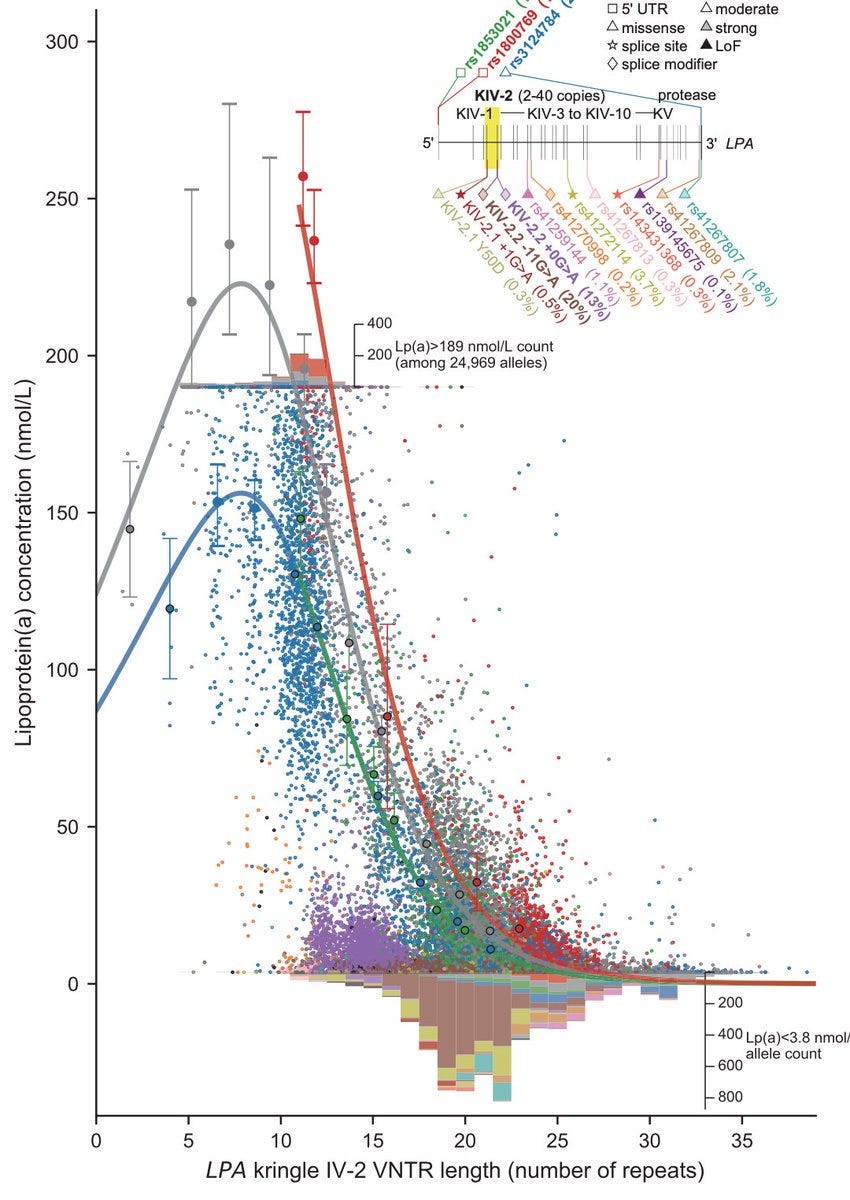
While the linear relationship between LPA VNTR repeats and Lp(a) levels was well-known, here the authors went a step further to reveal the "non-linear and cis-epistatic effects" of LPA variants on Lp(a) levels.
As the number of repeats drops, the Lp(a) levels rise; this relationship weakens at around 12 repeats and breaks and inverts at 8 repeats. The relationship curve is further modified by other missense variants suggesting epistatic effects.
When these non-linear and epistatic effects are taken into account, the variance explained by LPA variants rises from a previously estimated ~60% to a whopping ~90%, which is almost the twin h2 (93%) of Lp(a)--perhaps the first solved case of missing heritability.
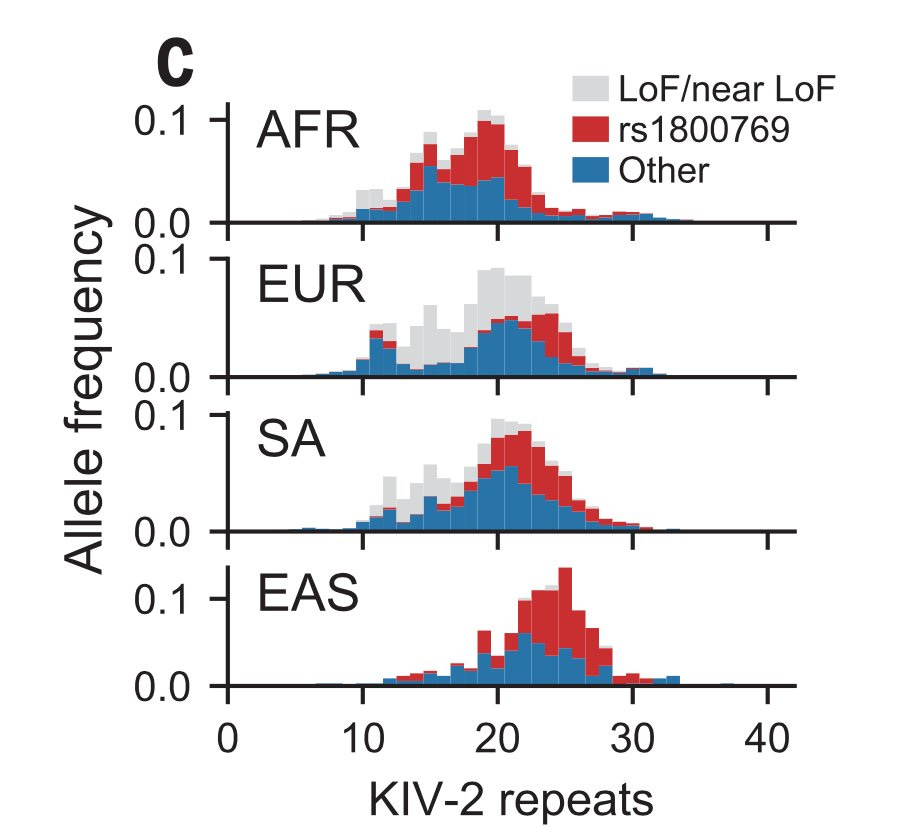
The LPA genetics also offers some great insights into the phenotypic and genetic differences across populations. For a trait such as Lp(a) whose variations are almost completely genetically determined, what would be the reason that the genetic prediction works well in pop A (e.g. Europeans) and poorly in pop B (e.g. Africans)?
The answer is obviously that the genetic prediction model doesn't include all the genetic variants from both populations A and B. The model has missed important variants that are likely more frequent in pop B than in A.
The Lp(a) levels are almost fourfold higher in Africans than in Europeans. But genetically predicted Lp(a) levels using VNTR alone do not capture this 4x difference between Europeans and Africans—a sign that an important piece of information is missing in the prediction model.
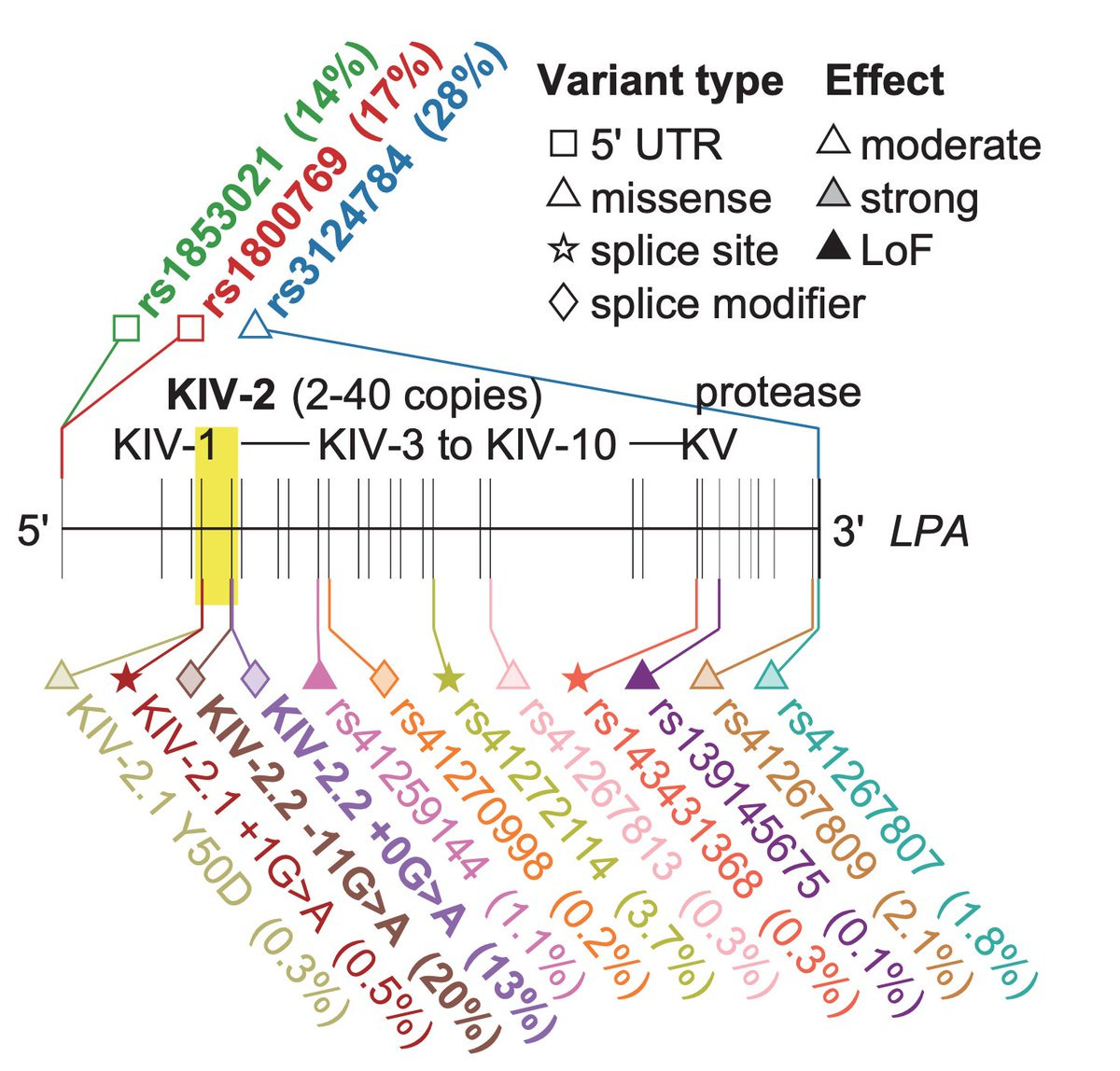
The authors found that certain Lp(a) increasing variants such as rs1800769, a 5' UTR variant, are more frequent in Africans than Europeans (MAF 46% vs 17%) and predominantly drive the between pop differences.
Incorporating such variants in the genetic prediction model closes the gap between the predicted and measured Lp(a) levels (blue and orange points in the image below).
This is a great demonstration of why it's crucial to study diverse populations and why it's crucial that we understand the between-population genetic differences before introducing genetic tests in the clinic. Imagine if an Lp(a) genetic test using VNTR alone is used to screen individuals to identify those at risk for CAD, most of the Africans who are truly at high risk will be missed.
Finally, LPA variants are an excellent example of an allelic series. Note that the different VNTR repeats themselves contribute to allelic series as they lie on distinct haplotypes.
Allelic series are interesting and also useful for many reasons. They offer a natural way of inferring how different levels of genetic perturbations impact the phenotype. Observing an allelic series in a GWAS is a sign that the locus is biologically important and probably holds a potential drug target.
LPA is just one of the many fascinating stories that you'll find in this paper. But I'll stop here. Many congratulations to Po-Ru Loh and the team for this great piece of work.






At deCODE we have looked into Lp(a) attributed causing cardiovascular diseases and have reported it in 2019 in JACC
https://www.sciencedirect.com/science/article/pii/S0735109719380386?via%3Dihub
Patrick