Of Mice and Men
And Women—Human Discoveries That Derived Inspiration from Mice Research
Happy Weekend! Recently, I've been reading about the origin story of lab mice in an essay, The Mouse as a Microscope, by Alex Telford1.
As a human geneticist working in the drug development field, I, like many others, have often complained about mice being a poor animal model to study the complex human physiology. But I've also appreciated the big roles those small mammals have played in biomedical research, particularly in human genetics. A few days ago, I tweeted about a new discovery of a rare monogenic cause of childhood onset systemic lupus erythematosus (SLE)—gain of function mutations in UNC93B1, encoding a toll-like receptor (TLR)-trafficking protein—published in Nature Immunology. The discovery reminded me of another post from two years ago on the discovery of a gain of function mutation in TLR7 as a monogenic cause of childhood onset SLE. With Alex's story still fresh in my mind, an aspect of both the studies kept coming back to me: the roles lab mice have played in establishing the links of TLR7 and UNC93B1 with SLE.
SLE is an autoimmune disease. Individuals with SLE produce autoantibodies against their own DNA, commonly known as antinuclear antibodies (ANAs). Under normal physiological conditions, DNA never leaves its home, the nucleus. When it does, it causes trouble. It alarms the innate immune system. Humans have evolved by fighting against pathogens like viruses for thousands of years. Our immune system is pretty good at spotting a pathogen intruder. When a virus enters a human cell and start replicating, their nucleic acids are readily sensed by a special family of badass proteins, TLR7, TLR8 and TLR9, that live in the endosomes. Activation of TLRs leads to consequences such as cytokine storm, B cell activation etc. to fight against the intruder. In patients with SLE, there is constant activation of TLRs without any viral infection, either because of frequent release of nuclear DNA into the cytoplasm or abnormally sensitive TLRs.

Presence of ANAs is diagnostic of SLE, which is known for a long time. ANA test is highly sensitive but not specific for SLE. As Derek Lowe puts it in one of his In the Pipeline articles, "if you are negative on an ANA, you basically don't have lupus. But if you're positive, you could still have something else, because that's a common positive in a number of autoimmune diseases". The groundbreaking discoveries of the TLRs' role in pathogen nucleic acids sensing in the early 2000s gave the first clues on the link between TLRs and SLE. It biologically made sense that TLR over activation could cause SLE. But the real-world evidence wasn't there to prove this causal link, though there were some hints. You see, TLR7 and TLR8 are on the X chromosome, and SLE is more almost 10-times more common in females than males. Scientists always thought that these two facts are not unrelated. Is it possible that the increased prevalence of SLE in females could be due to increased expression of TLR7 in the immune cells? We know the answer to that question today, that certain immune cells in females escape the random X inactivation and double the dosage of their X chromosomal genes, including TLR7. But in the early 2000s, scientists weren't sure. They were just beginning to connect the dots.
What if we could stitch an extra copy of TLR7 into the Y chromosome, will that increase the risk of SLE in males? Well, it turned out that is what exactly happened when scientists crossed B6 female mice (B6 or C57BL/6 is the most commonly used inbred mouse strain) with SB/Le male mice (SB/Le is a special strain used to study autoimmune diseases). This resulted in a hybrid that showed extreme autoimmunity. The animals spontaneously developed a lupus-like disease. The reasearchers at the National Institute of Allergy and Infectious Diseases in NIH named their creation Yaa (Y-linked autoimmune accelerator). It appeared that the crossing of B6 and SB/Le strains resulted in some genome exchange that pushed the autoimmune risk of SB/Le male mice through the roof. There was a clue that X chromosome from the B6 strain has something to do with it. Because Yaa mice came only from an SB/Le dad and a B6 mom. But not the other way around.
When researchers took a closer look at the Yaa’s genome, they stumbled upon something wild: an uninvited guest, a tiny piece of X chromosome containing the TLR7 gene, lurking inside the Y chromosome. So, the Yaa mice have got two copies of TLR7: one of their own in the X and another in the Y as a result of a translocation. The TLR7 duplication amplified the already autoimmune susceptible genome derived from the SB/Le strain. This spontaneous mutation provided a causal evidence that TLR7 over activation results in SLE. Conversely, when TLR7 is deleted in lupus mice model, scientists found, the autoimmunity is dampened.

Despite the overwhelming lines of evidence, no strong human genetic findings have surfaced connecting TLR7 gain of function with SLE susceptibility for a long time. The first such report was in 2022, which I have summarized in a Twitter thread. Scanning through the genome of a 7 yr old girl with a refractory, early onset SLE, a research team in Australia (Brown et al. Nature 2022) caught a de novo missense variant in TLR7 within a highly conserved sequence that encodes the ligand-binding part of the TLR7 protein (TLR7’s ligands are single stranded RNAs). Introducing this variant in mice triggered autoimmunity. If you read the paper, you'll appreciate how this single variant has opened a window for the immunologists to peek into the world of a SLE pathogenesis, revealing tons of insights. But what fascinated me most is that the authors knew where to look at. Thanks to prior knowledge on the role of TLRs in SLE that came mainly from mice studies.
The memory of the TLR7's story is what made me excited when I came across the Nature Immunology paper by a research team in China (Al-Azab, Idiiatullina, et al.) on gain of function mutations in UNC93B1 causing childhood-onset SLE. The beauty of these discoveries is that they are all interconnected, and discovery of one gene exerts a domino effect on the other. If too much TLR7 causes SLE, other proteins upstream or downstream of TLR7 signaling should have genetic links to SLE. One such protein is ‘unc-93 homolog B1’ encoded by UNC93B1.
Until the early 2000s, UNC93B1 was one of those genes encoding a protein with a weird name and an unknown function. Then born a mouse with the ‘3d’ mutation. Both of its parents were created using gametes bathed in a chemical called N-ethyl-N-nitrosourea, a powerful mutagen2. One of the hundreds of random chemically induced mutations hit UNC93B1, resulting in an immunodeficiency phenotype. It turned out that one stone killed three birds—TLR3, TLR7 and TLR9, opening the doors to all sorts of pathogens. The immunologists at The Scripps Research Institute who discovered this UNC93B1 mutation decided to call it ‘3d’ as it caused 3 defects in the immune system. Studying the ‘3d’ homozygous mice revealed the critical role unc-93 homolog B1 played inside the immune cells. It trafficked the TLRs from the endoplasmic reticulum to their site of action, the endosomes. Complete loss of UNC93B1 resulted in a selective immunodeficiency against intracellular pathogens. In the same year, the human counterparts of ‘3d’ mice were encountered by a research team in Paris (Casrouge, Zhang, Eidenschenk, Jouanguy, et al. Science 2006): two French children born from of a consanguineous marriage who were knockouts for UNC93B1 contracted a herpes simplex virus 1 infection in their brains.
Since its discovery, UNC93B1 has been extensively studied in the immunology field in the context of TLR signalling. In vitro mutagenesis screen has shown that gain of function mutations in UNC93B1 can amplify TLR signaling. However, no naturally occurring gain of function UNC93B1 mutations in humans were known. But it was only a matter of time. To find such mutations, all one has to do is look for them in the right individuals, which is exactly what the research team at the Guangzhou Medical University in China did.
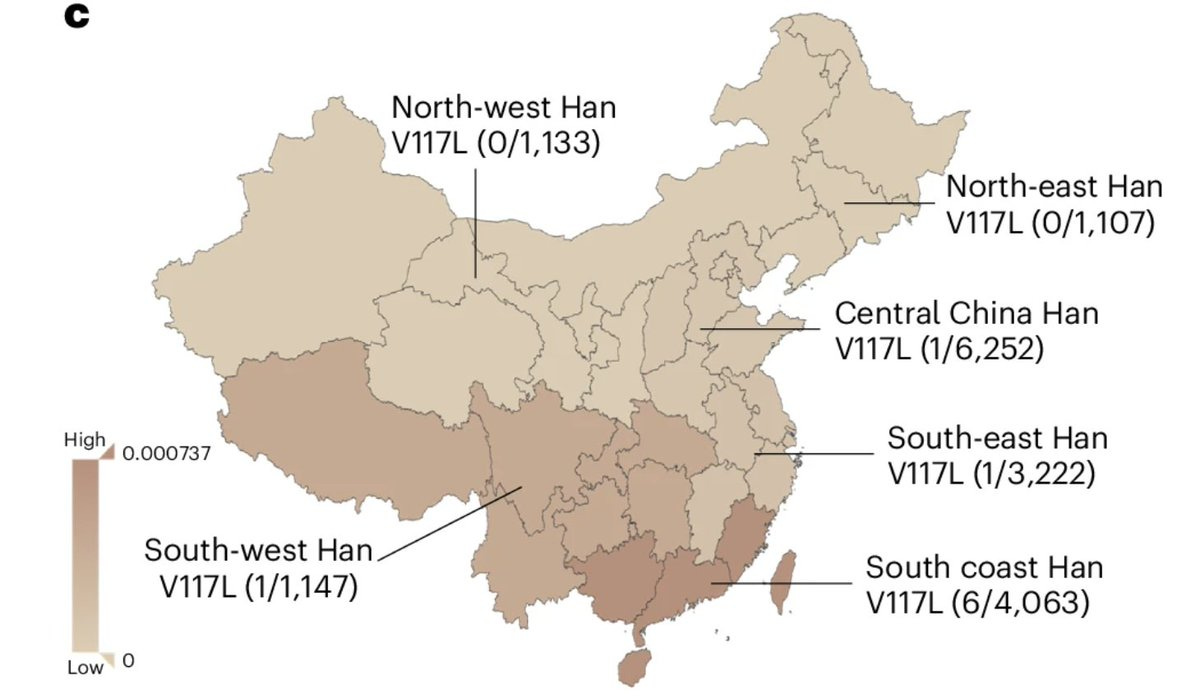
Scanning through the genomes of a childhood-onset SLE cohort, the team landed on the UC93B1 mutations, which they suspected to cause SLE. The mutations were in the part of the protein that binds specifically to TLR7. Introducing the mutation in mice triggered autoimmunity, confirming their intuition that these are gain of function in nature. The interesting part of the findings is that these mutations appear to be exclusive to East Asian ancestry. Particularly one of the mutations showed a gradient along the longitude with the highest frequency occuring in Southern coastal Han Chinese population. This is an important observation as this would mean that drug targeting TLR7 might be of value specifically to the patient communities from this part of the world, an example of personalized medicine in the context of ancestries. The good news is TLR7 antagonist do already exist that can be used to treat SLE in these patients.
To conclude, in both of the above stories, I wanted to emphasize the role lab mice played in kickstarting the research fields that led to an immense understanding of TLR signaling and how they are disrupted in SLE. If you crave for more inspiration about lab mice, I recommend Alex’s essay The Mouse as a Microscope.
Alex is a great writer and thinker. He has blogged essays on topics related to biotechnology and drug discovery.
One way to generate random mutations across the genome is to expose the animals to high dosage of chemicals, which will create hundreds or even thousands of de novo mutations in the germline. We can’t do this in humans deliberately. But we can leverage natural experiments. A remarkable study in Nature by Matt Hurles’ team at the Wellcome Sanger lnstitute in UK (Kaplanis et al.) has found that one of the rare causes of hypermutation events in humans is paternal exposure to chemotherapeutic agents just before conception.




