The underappreciated role of rare disease in drug development
Happy Friday! I’ve been thinking about the role of rare diseases in drug development this week. Drug companies are racing to sequence healthy volunteers in millions. But very few care about rare disease patients. As much as a healthy human carrying a beneficial mutation helps with drug development, a rare disease patient carrying a pathogenic mutation equally provide useful information. Rare disease informing drug development is one of my favorite topics. Two striking examples that I have highlighted before on Twitter (and on The Genetics Podcast) include the patients with neurodevelopmental disorder patients with BCL11A haploinsufficiency informing about the efficacy of BCL11A knockdown in blood cells to treat sickle cell disease and patients with a rare muscle disease caused by recessive mutations in HMGCR informing about a common adverse effect of statins. Falling under this theme is also the story below that I wrote on Twitter two years ago, which I chose for this week’s post.
From the Twitter archives
A functional study of six de novo missense variants in KCKN3 identified in 9 probands from the Deciphering Developmental Disorders (DDD) cohort highlights the importance of a K+ channel in the pathophysiology of sleep apnea.
This is a great example and a reminder that our biological understanding of many of the common diseases (in this case, sleep apnea) comes from rare monogenic disorders.
In this study, the authors report a new developmental disorder associated with sleep apnea caused by de novo gain of function missense mutations in a K+ channel coded by gene KCNK3.
Along with developmental delay, all the 9 probands had sleep apnea, a phenotype that is rarely (0.6%) seen in the entire DDD cohort (>13k), suggesting an extraordinary genotype-phenotype specificity.
One thing I love about this paper is that connecting the KCNK3 mutations with sleep apnea merges two streams of literature.
On one side, scientists were studying if we can use potassium channel blockers to dampen the mechanoreceptors in the upper airway muscles to treat sleep apnea. Here is a report in a pig model.
Although it was known that inhibitors (currently being evaluated in clinical trials for sleep apnea) are probably acting via the KCNK3 coded channels, no human genetics evidence existed to date to support that. And the so far known KCNK3 mutations were loss of function in nature, which had no links to sleep apnea but linked to an entirely different phenotype--pulmonary hypertension.
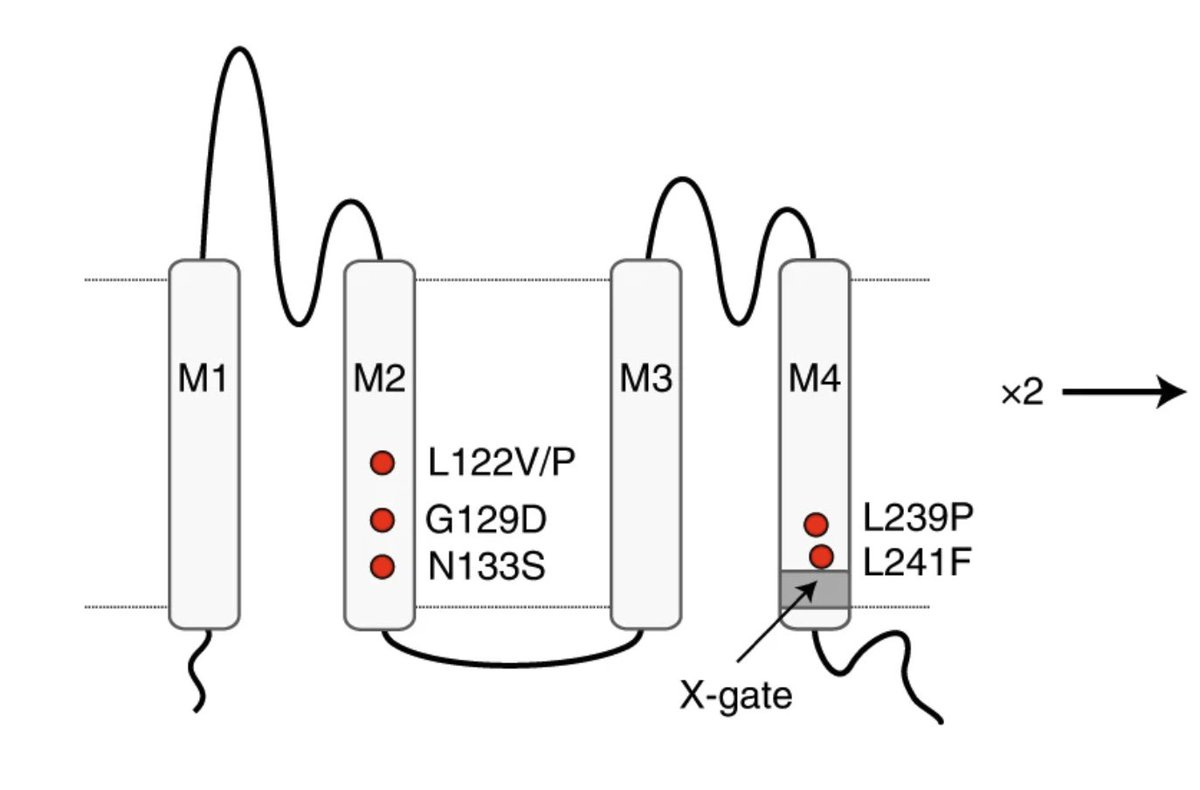
On the other side, scientists were studying how the potassium channels work by zooming in all the way into the crystal structure of the channel protein. Here is an amazing paper that reports the discovery of a second gate, called X gate, in a family of K+ channels to which KCNK3 belongs to.
Six amino acid residues 243-VLRFMT-248 in the M4 helices of two subunits of the dimeric protein crosses to create a second gate (X gate) and is essential for many channel ligands (anesthetics, neurotransmitters) to act.
In vitro experiments suggest that mutations surround this X gate lead to a gain of function phenotype, i.e. over activation of the channel due to failure of the X gate to close the main gate.
Given that inhibitors of this channel are hypothesized to treat sleep apnea, naturally occurring gain of function mutations in this channel should cause monogenic forms of sleep apnea. But no such mutations have been documented before. That critical piece of evidence now come from these 9 probands from different families who all got hit by de novo mutations very close to the X gate of KCNK3 channel. It just blows my mind to see such rare unfortunate mutation events leading to extraordinary biological insights.
Through functional studies, the authors confirm that all of these de novo missense mutations are indeed gain of function in nature. Fascinatingly, the mutations that lie right next to the X-gate in M4 helix have milder consequence. And the mutations that lie on the M2 helix which is away from the X gate in the linear space, but lie closer to X gate in the 3D space have severe consequence. The experimental results (channel activations) and clinical profile are in remarkable agreement.
The discovery of these mutations (results of nature's experiments in humans) finally validates the therapeutic hypothesis that K+ channel blockers inhibiting KCNK3 can treat sleep apnea. An amazing illustration of how human genetics complements drug development.
The paper also has a happy ending. There seem to exist highly potent inhibitors with extreme specificity to these channels that could be used to treat the sleep apnea in these patients. A rare instance where a genetic diagnosis is pointing to a promising treatment.